Search Count: 65
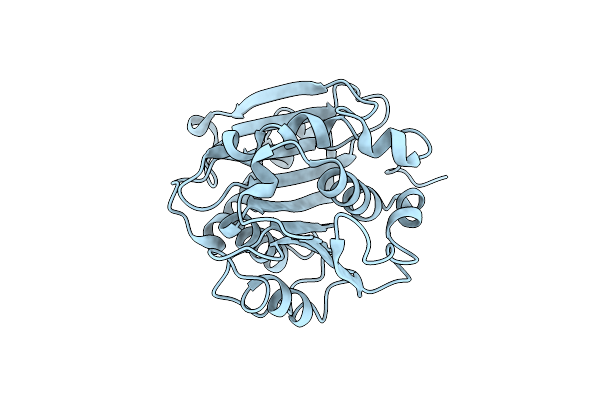 |
Crystal Structure Of A Novel Alpha/Beta Hydrolase From Thermomonospora Curvata In Apo Form
Organism: Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-12-27 Classification: HYDROLASE |
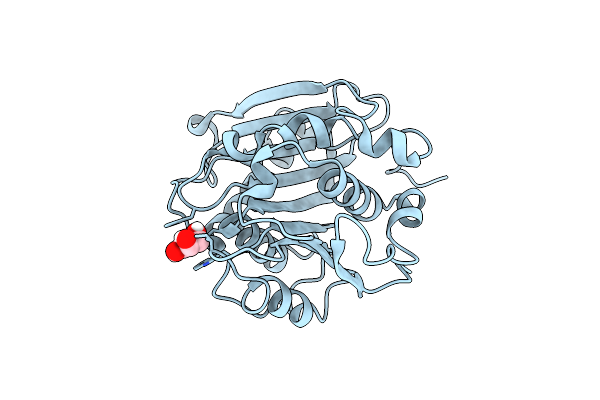 |
Crystal Structure Of A Novel Alpha/Beta Hydrolase From Thermomonospora Curvata With Glycerol
Organism: Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: GOL |
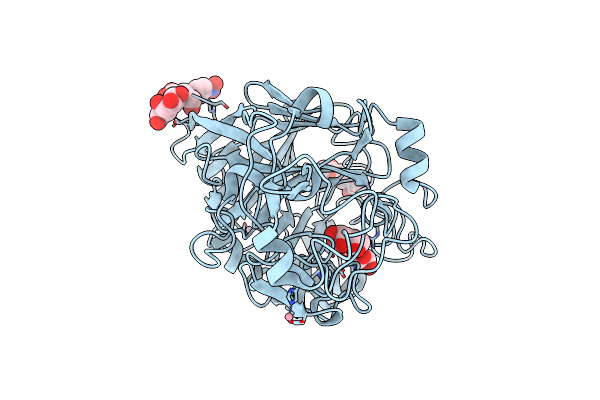 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-03-09 Classification: HYDROLASE Ligands: NAG, BGC, GAL, NPO, CO |
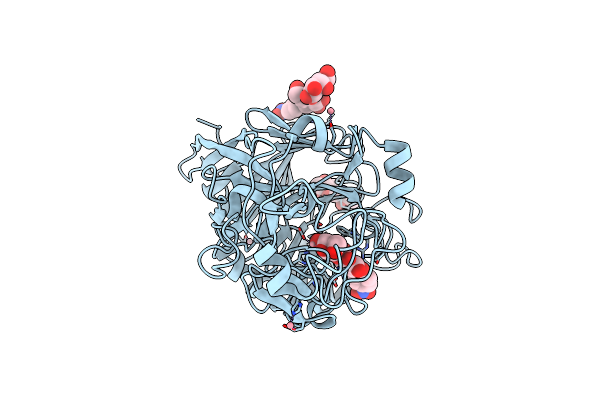 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-03-09 Classification: HYDROLASE Ligands: NAG, NPO, CO |
 |
Improved Ligand Discovery Using Micro-Beam Data Collection At The Edge Of Protein Crystals
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: ARG, NA, EDO |
 |
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2021-07-14 Classification: HYDROLASE |
 |
Organism: Hypocrea jecorina (strain qm6a)
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-11-06 Classification: HYDROLASE Ligands: CO, NAG |
 |
Crystal Structure Of Aspergillus Oryzae Catechol Oxidase Complexed With Resorcinol
Organism: Aspergillus oryzae (strain atcc 42149 / rib 40)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: CU, MAN, NAG, RCO, PEO |
 |
Organism: Aspergillus oryzae (strain atcc 42149 / rib 40)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-05-09 Classification: OXIDOREDUCTASE Ligands: CU, MAN, NAG, EDO, FLC, PER |
 |
Organism: Aspergillus oryzae (strain atcc 42149 / rib 40)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-05-09 Classification: OXIDOREDUCTASE Ligands: CU, NAG, MAN |
 |
Organism: Amorphotheca resinae
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2018-05-09 Classification: HYDROLASE Ligands: NAG |
 |
Structural Studies Of A Glycoside Hydrolase Family 3 Beta-Glucosidase From The Model Fungus Neurospora Crassa
Organism: Neurospora crassa or74a
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Trichoderma reesei qm6a
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-09-20 Classification: OXIDOREDUCTASE Ligands: NAG, MAN, CU, SO4 |
 |
Organism: Trichoderma reesei qm6a
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2017-09-20 Classification: OXIDOREDUCTASE Ligands: NAG, MAN, CU, SO4 |
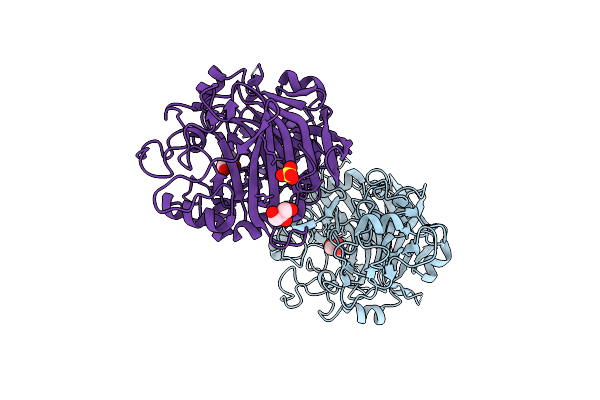 |
Radiation Damage To Gh7 Family Cellobiohydrolase From Daphnia Pulex: Dose (Dwd) 1.11 Mgy
Organism: Daphnia pulex
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, SO4 |
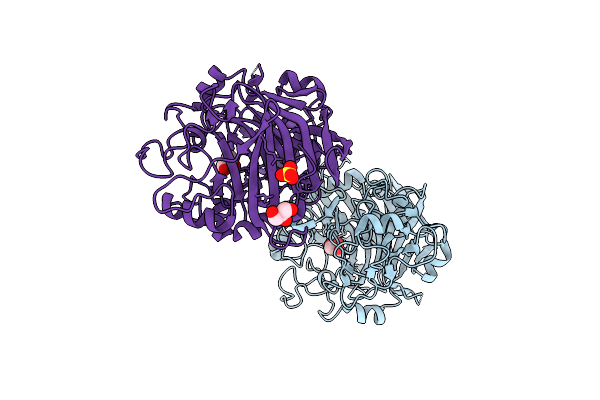 |
Radiation Damage To Gh7 Family Cellobiohydrolase From Daphnia Pulex: Dose (Dwd) 3.27 Mgy
Organism: Daphnia pulex
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, SO4 |
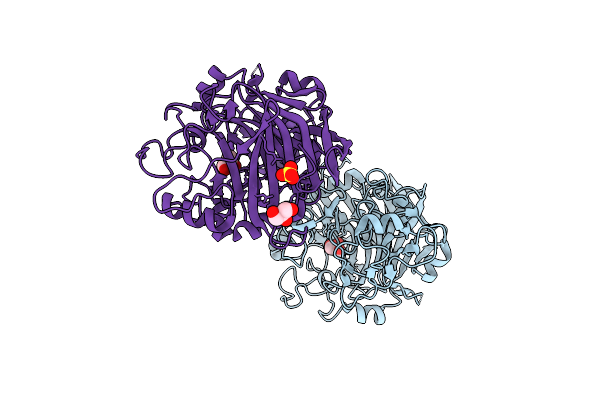 |
Radiation Damage To Gh7 Family Cellobiohydrolase From Daphnia Pulex: Dose (Dwd) 5.43 Mgy
Organism: Daphnia pulex
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, SO4 |
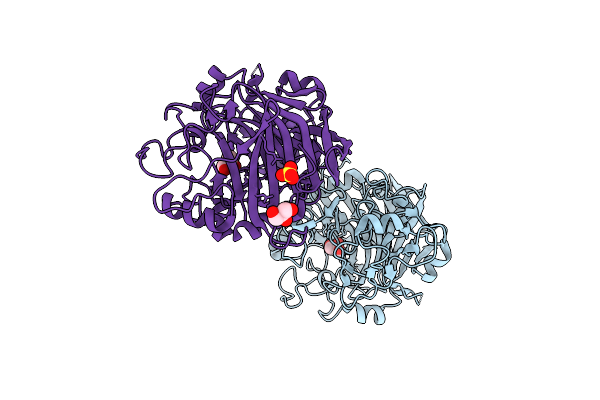 |
Radiation Damage To Gh7 Family Cellobiohydrolase From Daphnia Pulex: Dose (Dwd) 7.59 Mgy
Organism: Daphnia pulex
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Radiation Damage To Gh7 Family Cellobiohydrolase From Daphnia Pulex: Dose (Dwd) 9.75 Mgy
Organism: Daphnia pulex
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Radiation Damage To Gh7 Family Cellobiohydrolase From Daphnia Pulex: Dose (Dwd) 11.9 Mgy
Organism: Daphnia pulex
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, SO4 |

