Search Count: 46,148
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: DNA Ligands: LDP |
 |
Solution Structure Of An Intramolecular Anti-Parallel G-Quadruplex With A 5'-End Overhang.
|
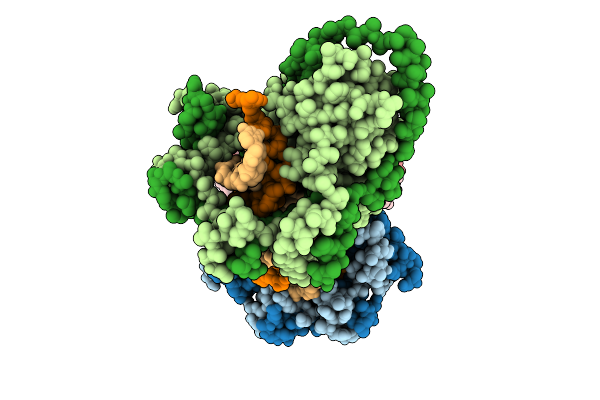 |
Organism: Mycobacterium tuberculosis, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
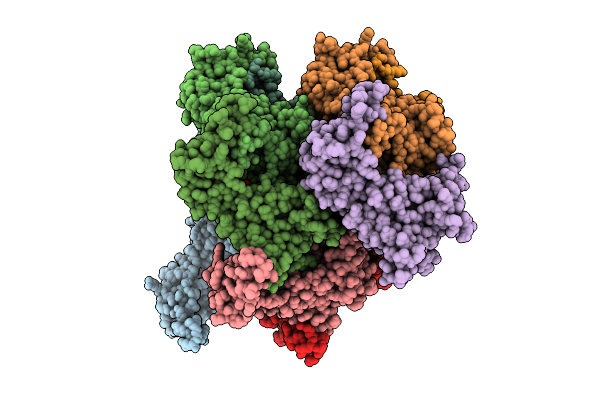 |
Eleven Polymer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutaion) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
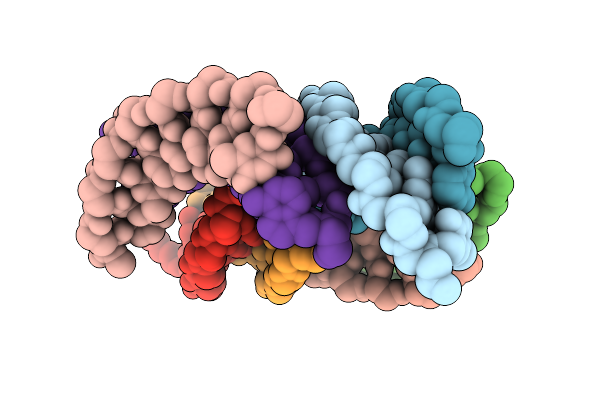 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PEPTIDE NUCLEIC ACID |
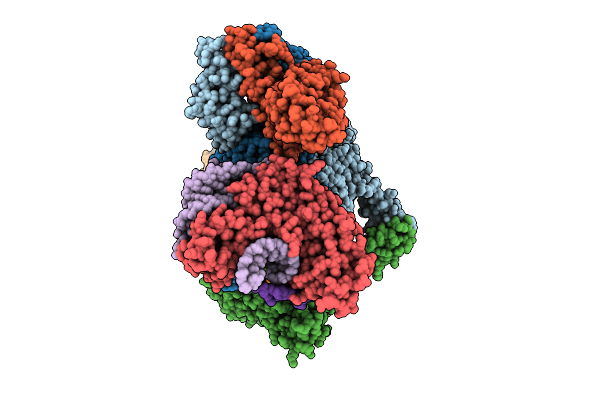 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ANTIVIRAL PROTEIN/DNA/RNA Ligands: ATP, ZN, MG |
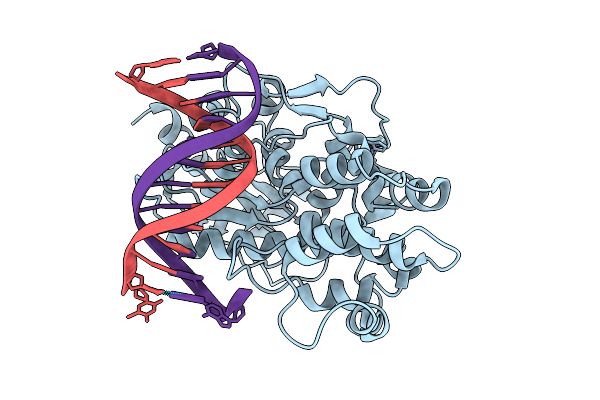 |
Crystal Structure Of Vanc21 In Complex With Its Target Dna (5-Bromouridine Substituted).
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
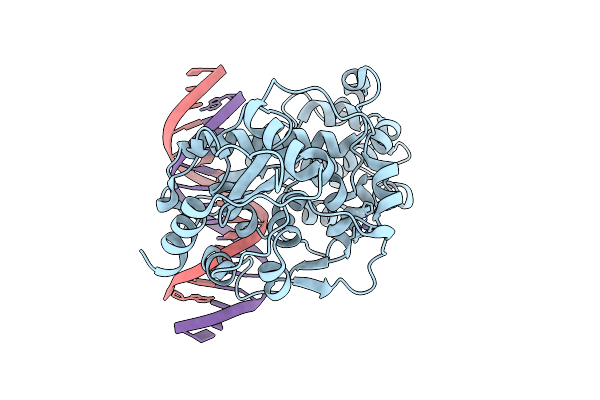 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
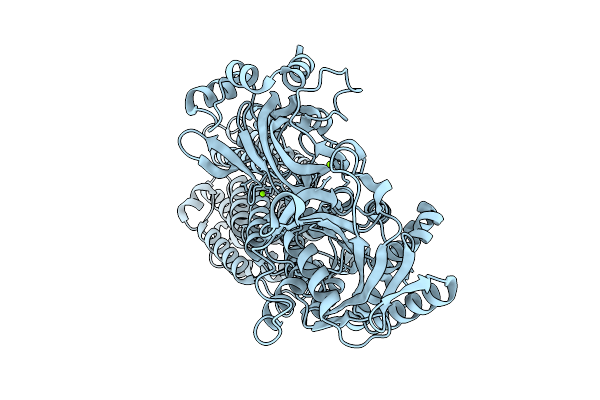 |
Organism: Lactococcus lactis subsp. lactis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: MG |
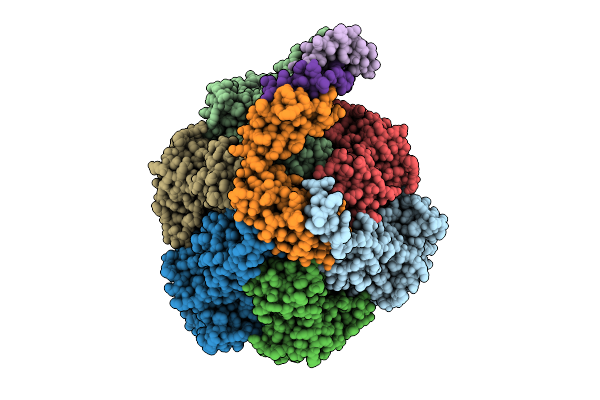 |
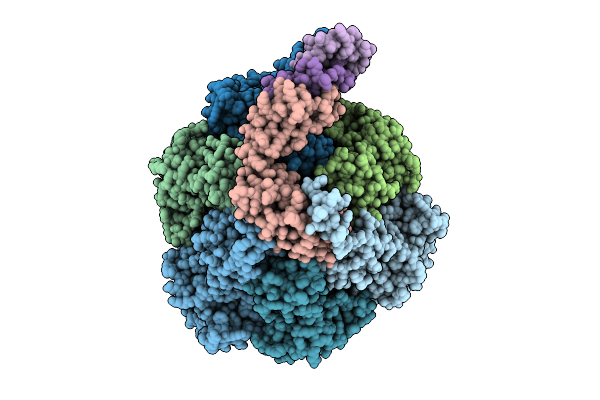
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, PO4 |
 |
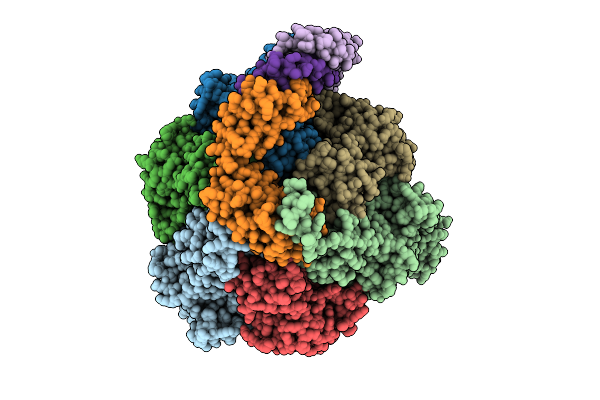
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
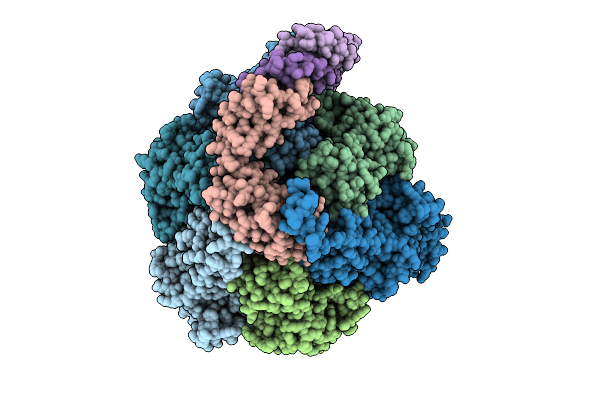
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
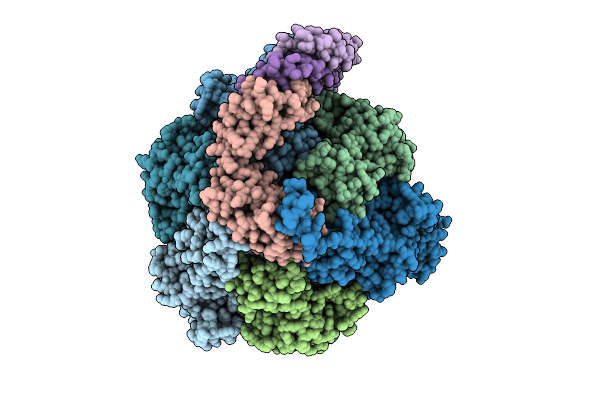
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |

