Search Count: 251
All
Selected
 |
Organism: Acanthamoeba polyphaga mimivirus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2026-02-25 Classification: TRANSLATION |
 |
Organism: Schizosaccharomyces pombe 972h-
Method: SOLUTION NMR Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN |
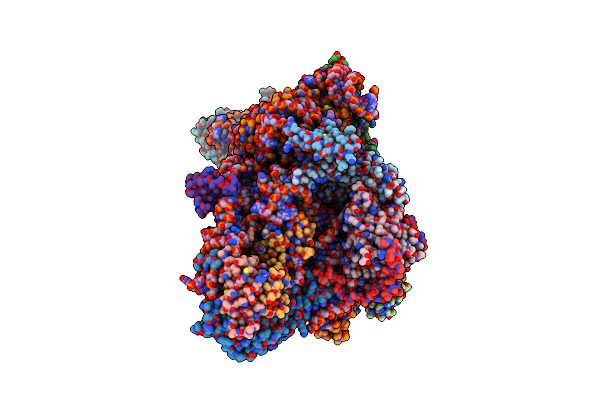 |
Sars-Cov-2 Nsp1 Bound To The Rhinolophus Lepidus 40S Ribosomal Subunit (Local Refinement Of The 40S Body)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens, Rhinolophus lepidus
Method: ELECTRON MICROSCOPY Resolution:2.10 Å Release Date: 2025-06-11 Classification: RIBOSOME Ligands: MG, K, ZN |
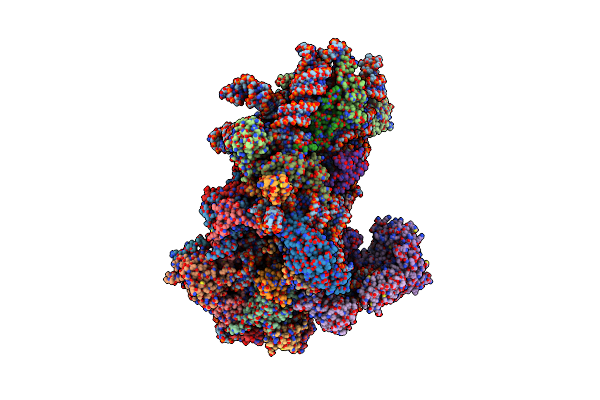 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Swivelled Conformation (Model Py48S-Auc-Swiv-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: 3HE |
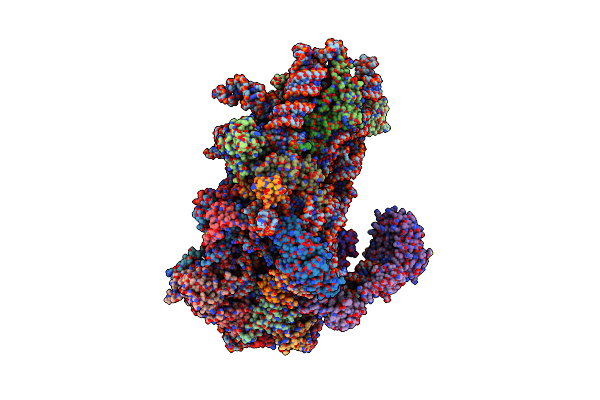 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
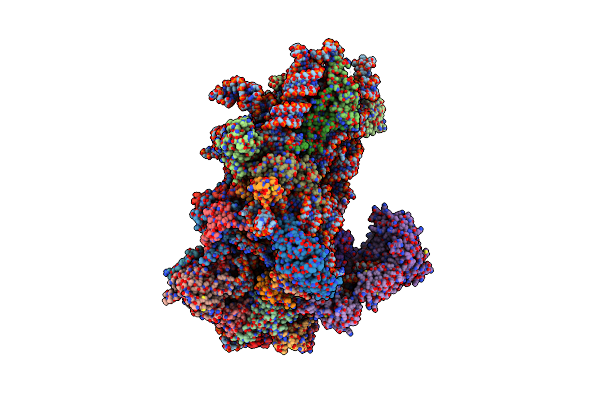 |
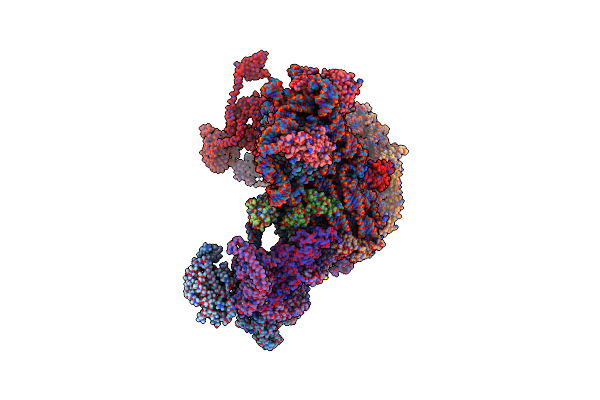
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
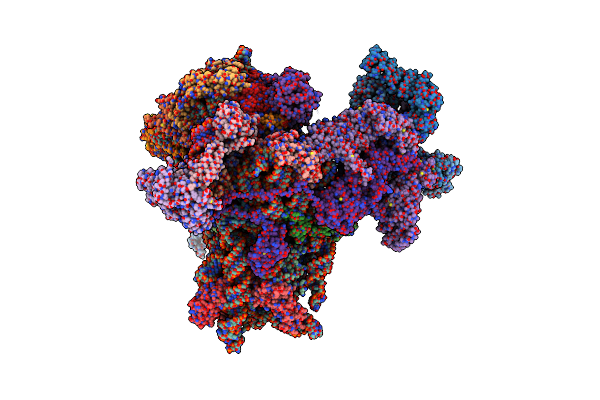
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: ANTITUMOR PROTEIN Ligands: MG, K, SPD, ZN, ACY |
 |
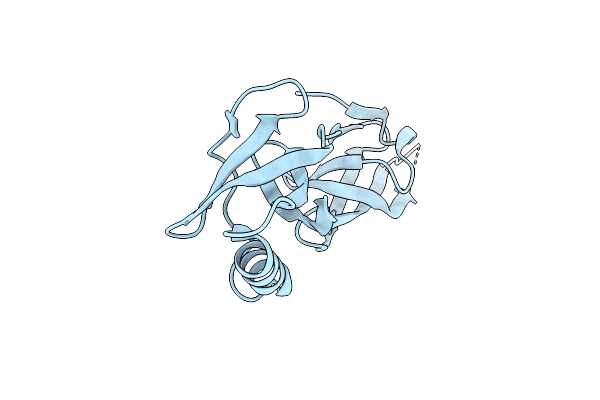
Structure Of Human 48S Translation Initiation Complex In Open Codon Scanning State (48S-1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RIBOSOME Ligands: MG, ZN, GTP, MET |
 |
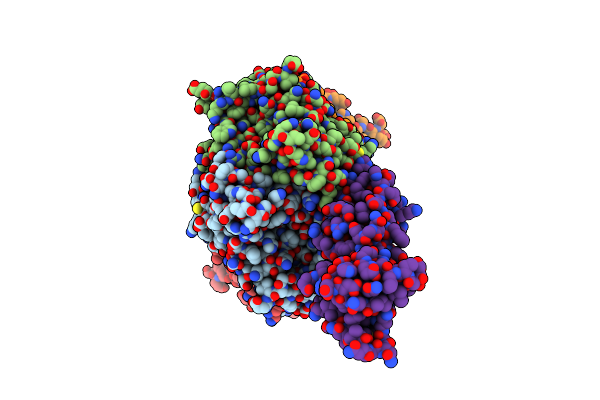
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-07-03 Classification: TRANSLATION |
 |
Organism: Trichomonas vaginalis
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: TRANSLATION Ligands: NAD, SPD |
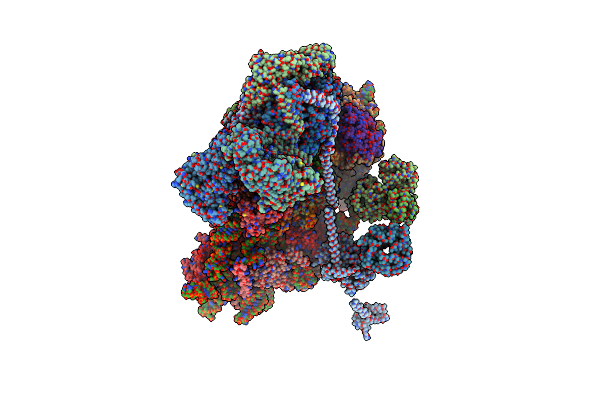 |
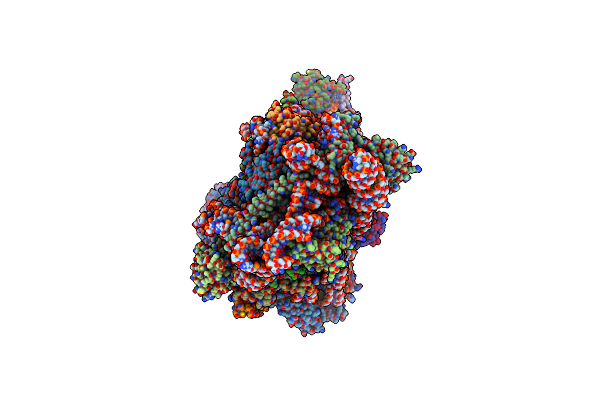
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RIBOSOME Ligands: MG, ZN |
 |
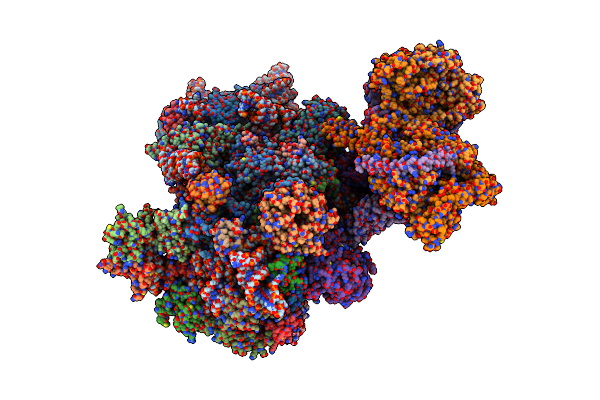
Organism: Homo sapiens, Bat hp-betacoronavirus/zhejiang2013
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: TRANSLATION Ligands: UNX, MG, ZN |
 |
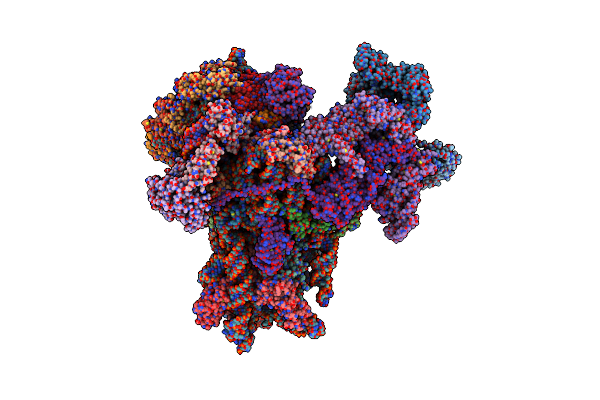
Organism: Middle east respiratory syndrome-related coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: TRANSLATION Ligands: ZN, GTP, MG, MET, UNX |
 |
Structure Of The Human 48S Initiation Complex In Open State (H48S Aug Open)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-11 Classification: RIBOSOME Ligands: ZN, MG |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: MN, AKG, EDO, MG |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: MN, SIN, EDO, MG |
 |
Drosophila Melanogaster Jmjd7 (Dmjmjd7) In Complex With Mn And N-Oxalylglycine (Nog)
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: MN, OGA, EDO, SO4 |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: MN, PEG, EDO, PD2 |
 |
Drosophila Melanogaster Jmjd7 (Dmjmjd7) In Complex With Mn And N-Oxalyl-D-Alanine (Noda)
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: MN, I78, EDO, ACT |

