Search Count: 10,895
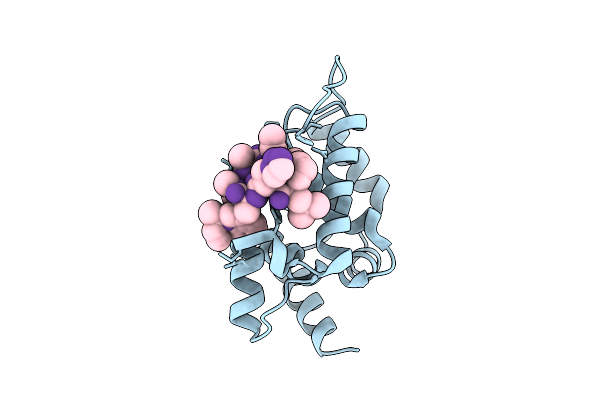 |
M. Tuberculosis Clpc1-Ntd Complexed With A Click Chemistry Analog Of Rufomycin
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIBIOTIC |
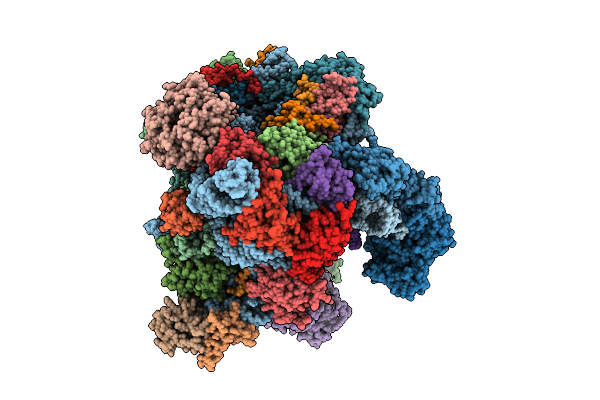 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
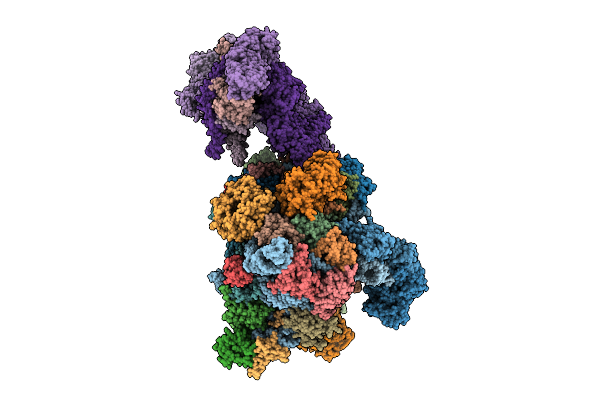 |
Late-Stage 48S Initiation Complex With Eif3 (Ls48S-Eif3 Ic) Guided By The Trans-Rna
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
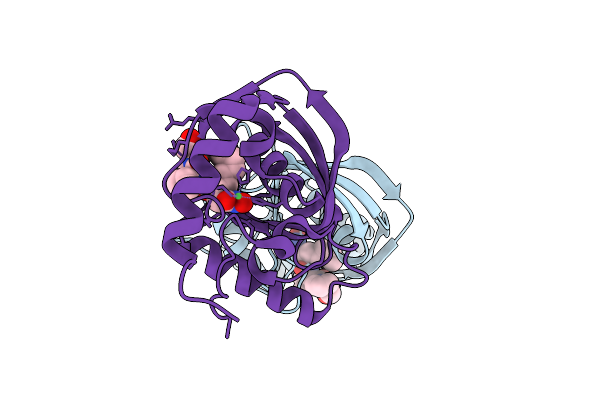 |
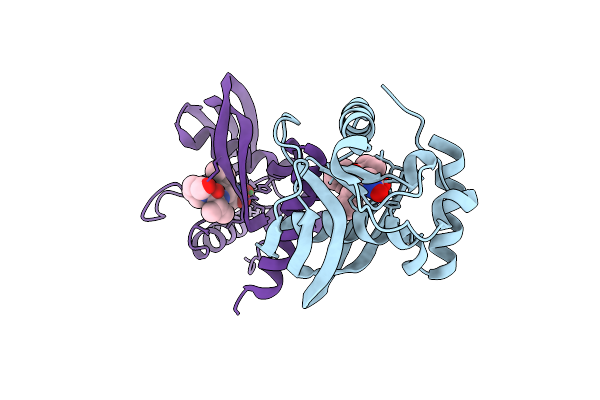
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NI, BB2 |
 |
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: BB2, NI |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PROTEIN FIBRIL |
 |
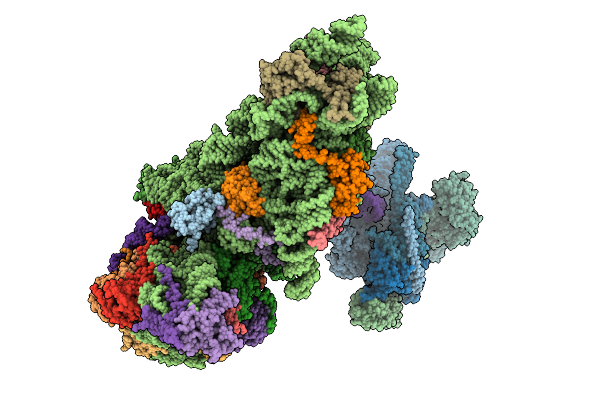
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
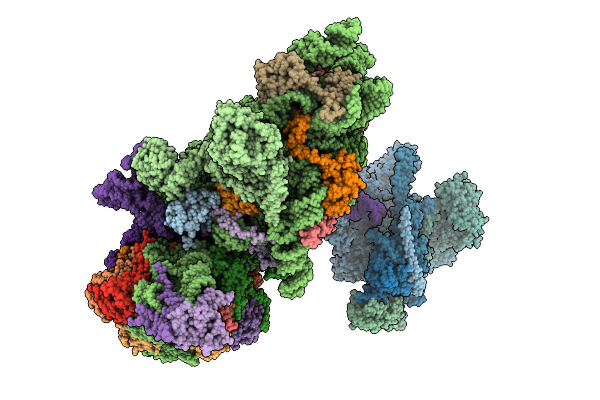
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
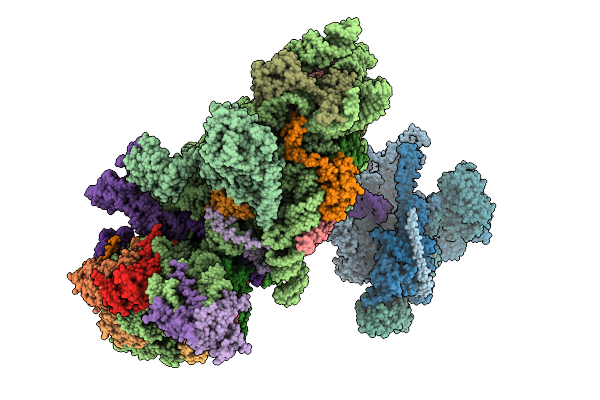
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
 |
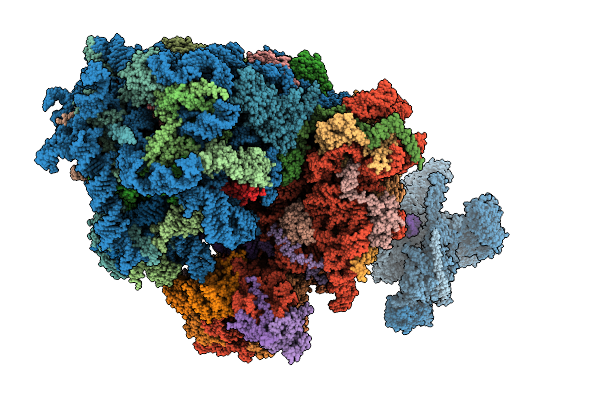
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
 |
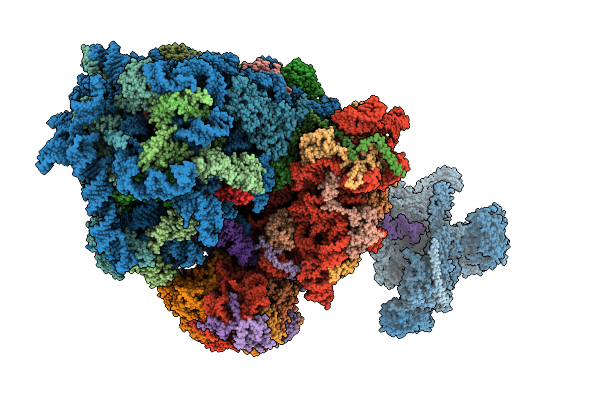
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
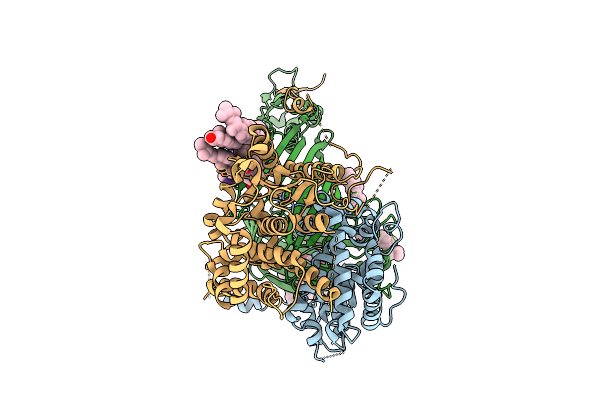 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
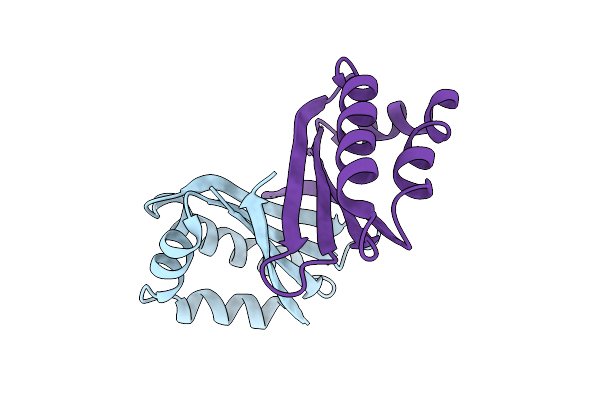 |
An Antibiotic Biosynthesis Monooxygenase Family Protein From Streptomyces Sp. Ma37
Organism: Streptomyces sp. ma37
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
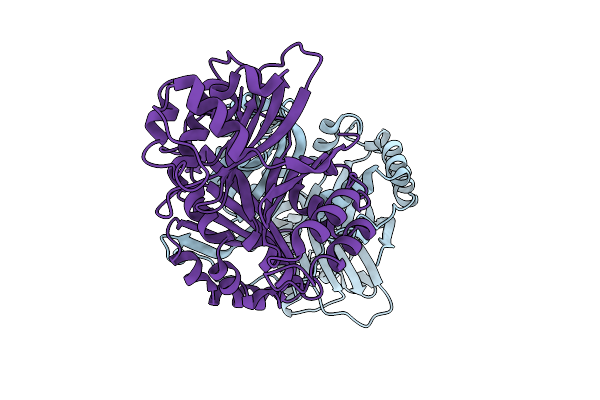 |
Crystal Structure Of A Polyketide Abm/Scha-Like Domain-Containing Protein Whie-Orfi From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |

