Search Count: 213
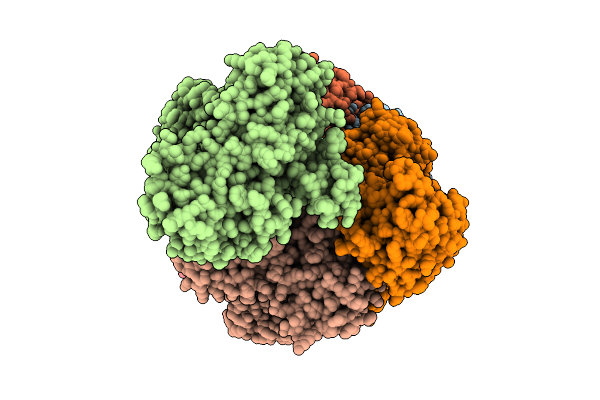 |
Cryoem Structure Of A Filamentous Soluble Pyridine Nucleotide Transhydrogenase
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: FLAVOPROTEIN Ligands: FAD |
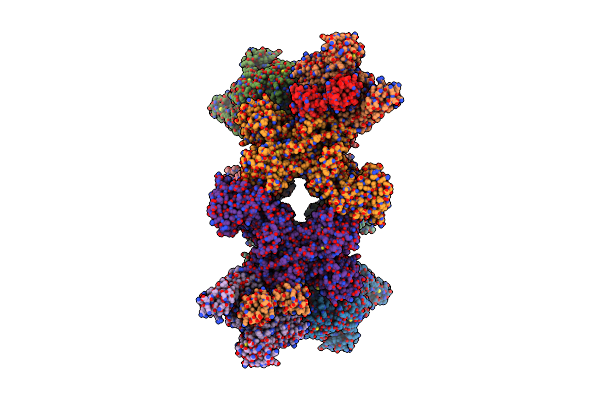 |
Cryo-Em Structure Of The Electron Bifurcating Transhydrogenase Stnabc Complex From Sporomusa Ovata (State 2)
Organism: Sporomusa ovata dsm 2662
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: ELECTRON TRANSPORT Ligands: FES, ZN, FMN, SF4, NAD, FAD, NDP |
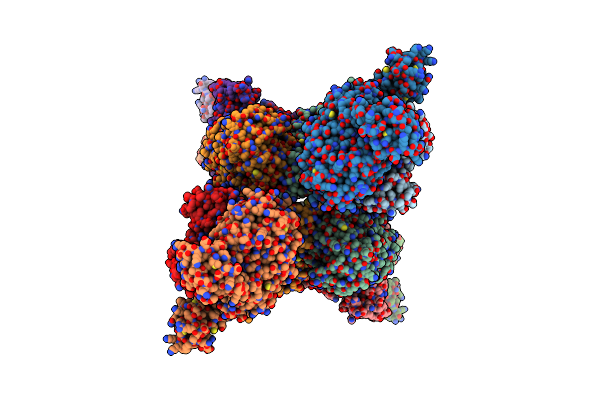 |
Cryo-Em Structure Of The Electron Bifurcating Transhydrogenase Stnabc Complex From Sporomusa Ovata (State 1)
Organism: Sporomusa ovata dsm 2662
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: ELECTRON TRANSPORT Ligands: FES, ZN, SF4, FAD |
 |
Structure Of Ovine Transhydrogenase In The Presence Of Nadp+ In A "Double Face-Down" Conformation
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: NAP, NAD, PC1, D12 |
 |
Structure Of Ovine Transhydrogenase In The Presence Of Nadp+ In A "Single Face-Down" Conformation
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: NAP, NAD |
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: MEMBRANE PROTEIN Ligands: PC1 |
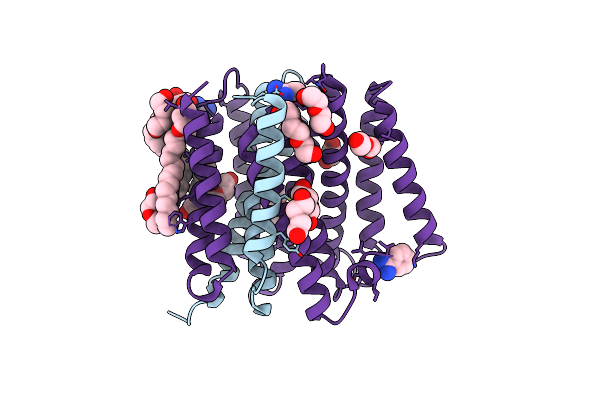 |
Critical Role Of Water Molecules For Proton Translocation Of The Membrane-Bound Transhydrogenase
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-05-10 Classification: OXIDOREDUCTASE Ligands: 1PE, PEG, OLC, BEN |
 |
Mechanism Of Transhydrogenase Coupling Proton Translocation And Hydride Transfer
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:6.93 Å Release Date: 2015-01-28 Classification: MEMBRANE PROTEIN Ligands: NAP, NAD |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2014-12-31 Classification: MEMBRANE PROTEIN Ligands: HG |
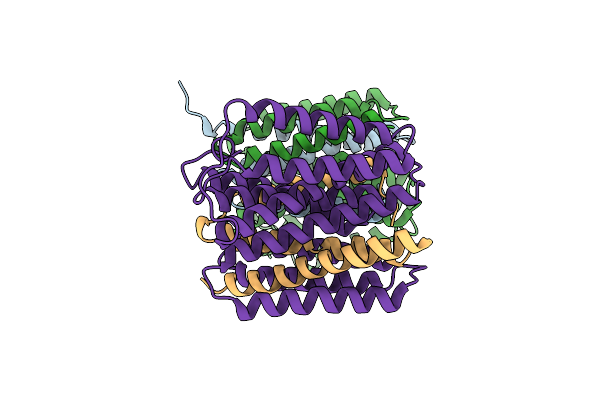 |
Crystal Structure Of Thermus Thermophilis Transhydrogeanse Domain Ii Dimer Semet Derivative
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2014-06-11 Classification: MEMBRANE PROTEIN |
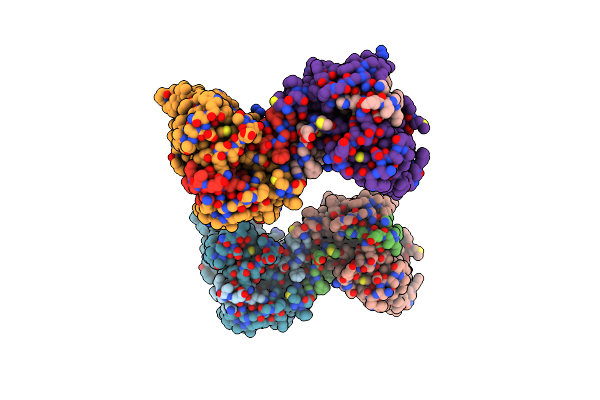 |
Mechanism Of Transhydrogenase Coupling Proton Translocation And Hydride Transfer
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2014-06-11 Classification: MEMBRANE PROTEIN |
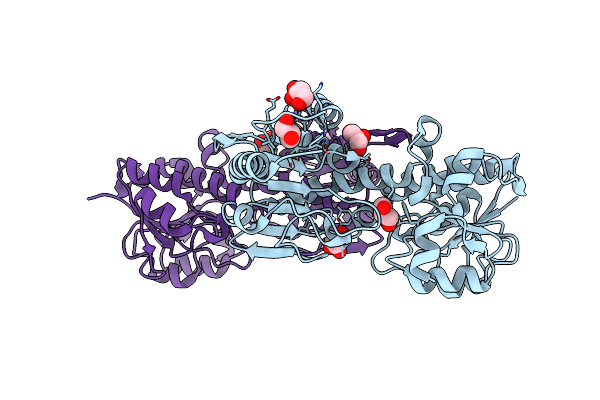 |
Crystal Structure Of The Alpha1 Dimer Of Thermus Thermophilus Transhydrogenase In P4(3)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE Ligands: GOL |
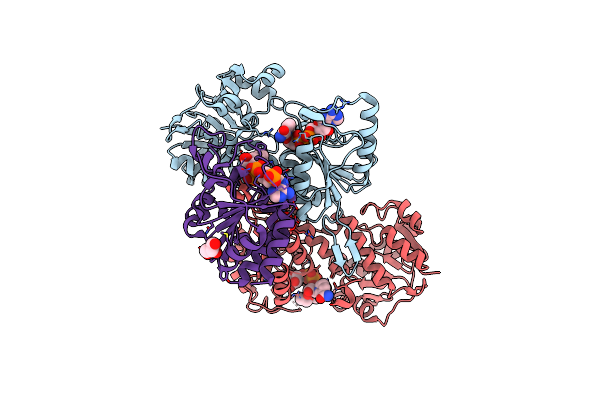 |
Crystal Structure Of Thermus Thermophilus Transhydrogenase Heterotrimeric Complex Of The Alpha1 Subunit Dimer With The Nadp Binding Domain (Domain Iii) Of The Beta Subunit
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE Ligands: NAD, CL, PGE, GOL, NAP |
 |
Crystal Structure Of Thermus Thermophilus Transhydrogenase Heterotrimeric Complex Of The Alpha1 Subunit Dimer With The Nadp Binding Domain (Domain Iii) Of The Beta Subunit In P2(1)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE Ligands: GOL, NAD, NAP |
 |
Crystal Structure Of The Alpha1 Dimer Of Thermus Thermophilus Transhydrogenase In P6
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-29 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-03-21 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Alanine Dehydrogenase/Pyridine Nucleotide Transhydrogenase From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2010-10-13 Classification: OXIDOREDUCTASE Ligands: CA, NA, EDO |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-03-13 Classification: OXIDOREDUCTASE Ligands: SO4, TXD, NAP |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2007-03-13 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, TXP |
 |
Complex Of The Domain I And Domain Iii Of Escherichia Coli Transhydrogenase
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2006-08-29 Classification: OXIDOREDUCTASE Ligands: NAD, NAP |

