Search Count: 1,18,288
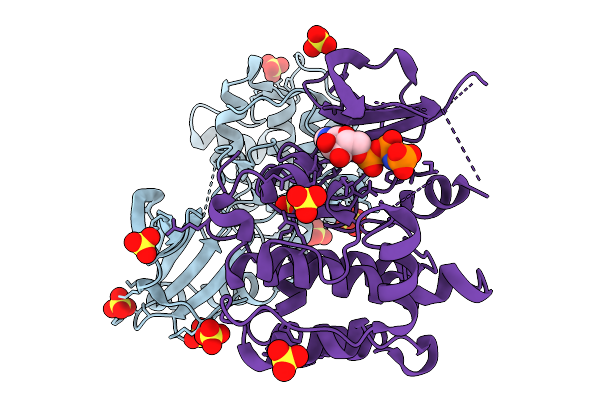 |
Crystal Structure Of Serine/Threonine-Protein Kinase (Aek1) T376D, S395D Mutant From Trypanosoma Brucei (Amp-Pnp)
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: SO4, ANP |
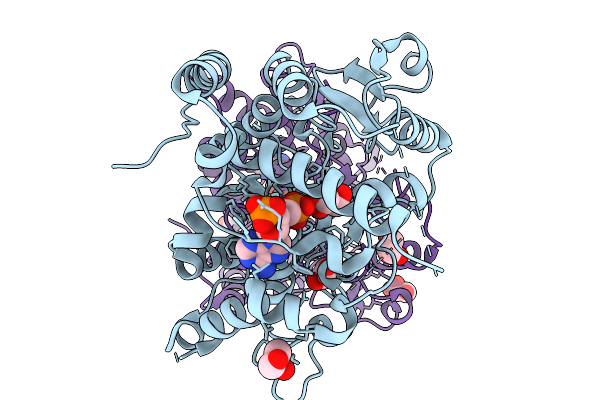 |
Crystal Structure Of Sorghum Sulfotransferase Lgs1 Reveals Sulfation-Assisted Bc-Ring Formation In Strigolactone Biosynthesis
Organism: Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: A3P, GOL |
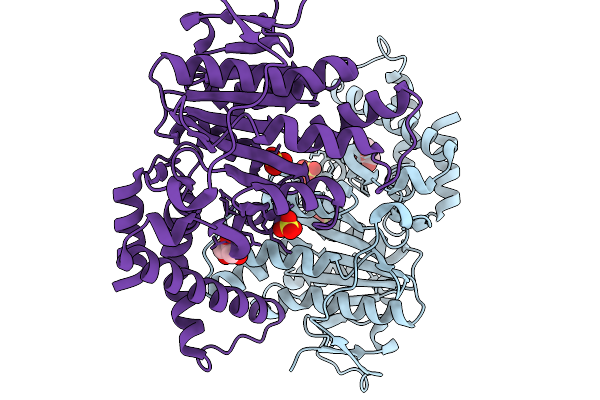 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: SO4, GOL |
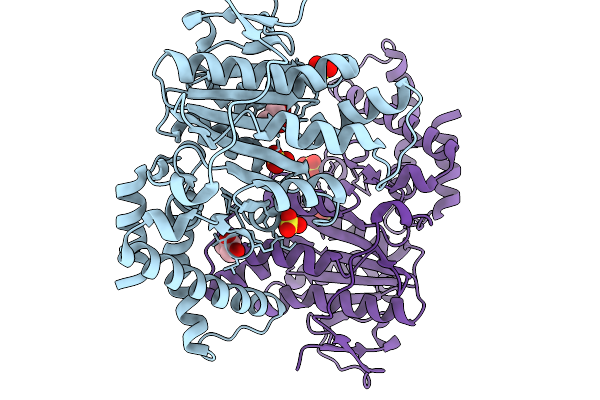 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: SO4, GOL |
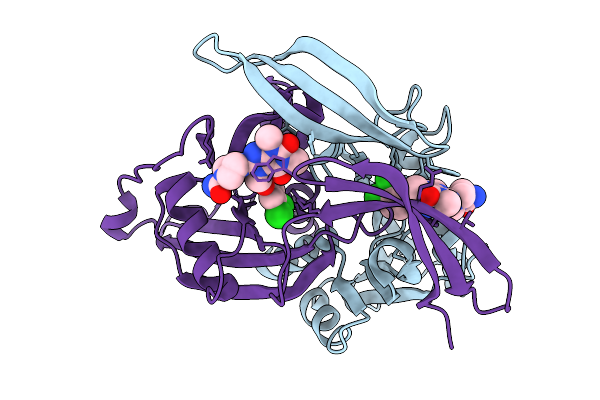 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: A1IIY, MG |
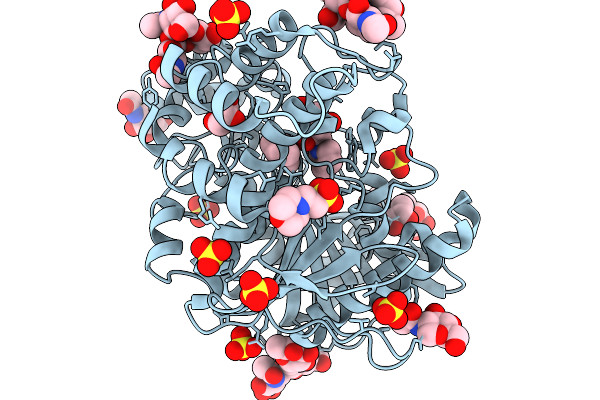 |
Structure Of Carbamoylated Recombinant Human Butyrylcholinesterase By The Biscarbamte 5-(1-Hydroxy-2-(Piperidin-1-Yl)Ethyl)-1,3-Phenylene Bis(Piperidine-1-Carboxylate)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: NAG, MES, GOL, A1I0D, A1I0F, SO4 |
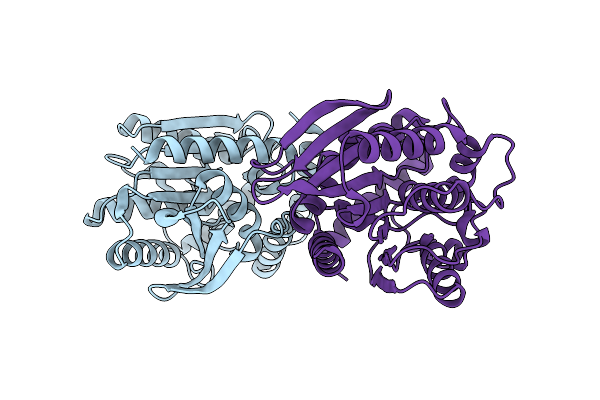 |
Crystal Structure Of A Pu Hydrolysis Enzyme Aes72 From Comamonas Acidovorans
Organism: Delftia acidovorans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-02-04 Classification: HYDROLASE |
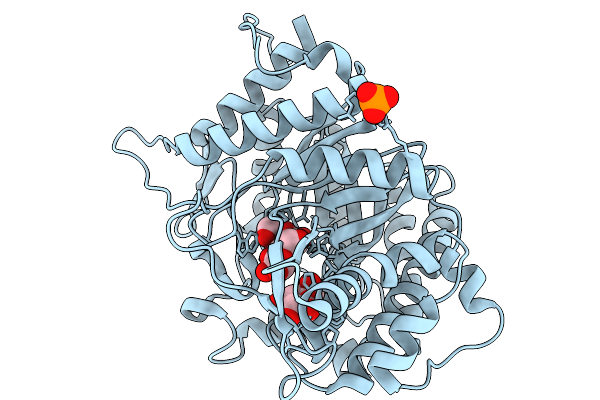 |
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4 |
 |
Crystal Structure Of Glucose Tolerant Beta-Glucosidase Unbgl1 Mutant (C188V)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: CL, PO4 |
 |
Crystal Structure Of Glucsoe Bound Glucose Tolerant Gh1 Beta-Glucosidase Mutant (Unbgl1_C188V)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4 |
 |
Crystal Structure Of Glucose Tolerant Gh1 Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: PO4, GOL, CL |
 |
Crystal Structure Of Glucose Bound Gh1 Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, GOL, PO4 |
 |
Crystal Structure Of Cellobiose And Glucose Bound Glucose Toleranant Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4, CL |
 |
High Resolution Crystal Strucutre Of Highly Glucose Tolerant Gh1 Beta-Glucosidase (Unbgl1_C188V_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of Glucose Bound Highly Glucose Tolerant Gh1 Beta-Glcosidase Mutant (Unbgl1_C188V_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4 |
 |
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: GS1, PO4, NA |
 |
Crystal Structure Of Glucose Bound Covalent Intermediate Of Gh1 Beta-Glucosidase (Unbgl1)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4 |
 |
High Resolution Crystal Structure Of Gh1 Beta-Glucosidase From Soil Metagenome (Unbgl1)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: PO4, CL, GOL |
 |
Crystal Structure Of Substrate Bound Gh5_22 Exo-Beta-Xylosidase From The Seaweed-Derived Thermophile Geobacillus Thermodenitrificans Os27
Organism: Geobacillus thermodenitrificans
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: GOL, FE |
 |
Crystal Structure Of Gh5_22 Exo-Beta-Xylosidase From The Seaweed-Derived Thermophile Geobacillus Thermodenitrificans Os27
Organism: Geobacillus thermodenitrificans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: FE |

