Search Count: 1,14,959
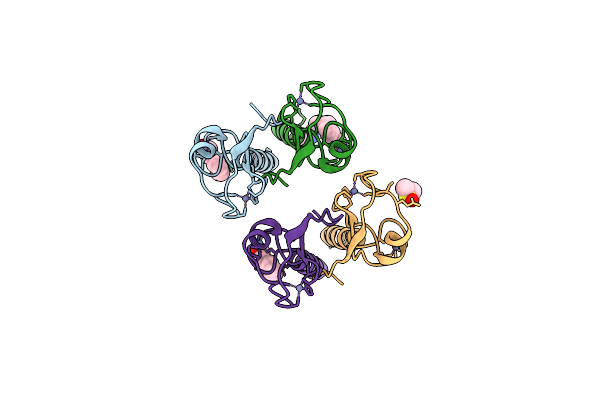 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1118527729
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, A1CX8, DMS |
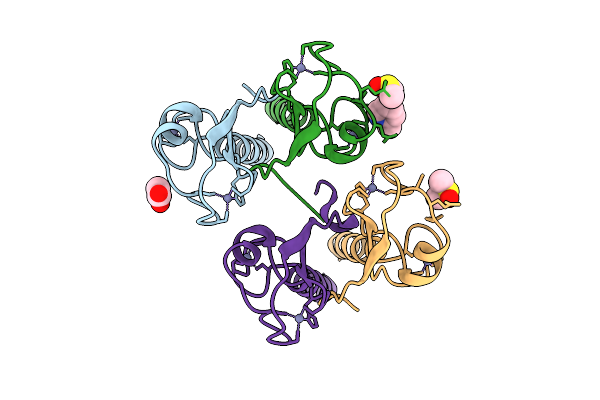 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1251361039
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, ACT, A1CYA, DMS |
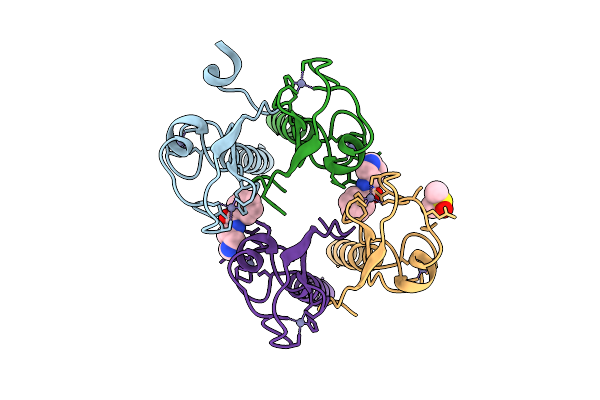 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1456069604
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, KF8, DMS |
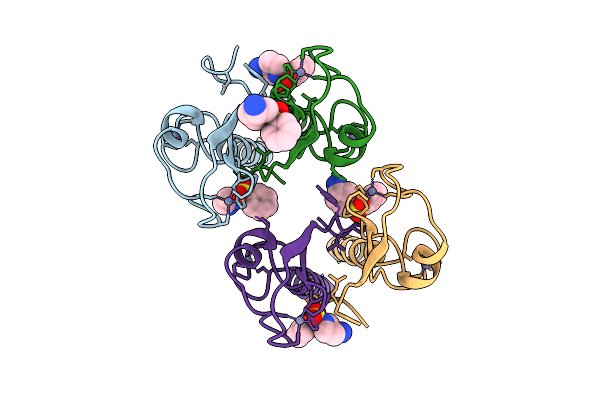 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1491353358
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, WNM |
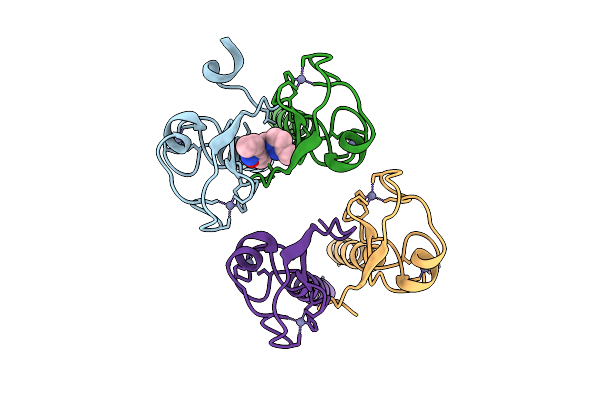 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1516316257
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, A1CYB |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1587220559
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, T1J |
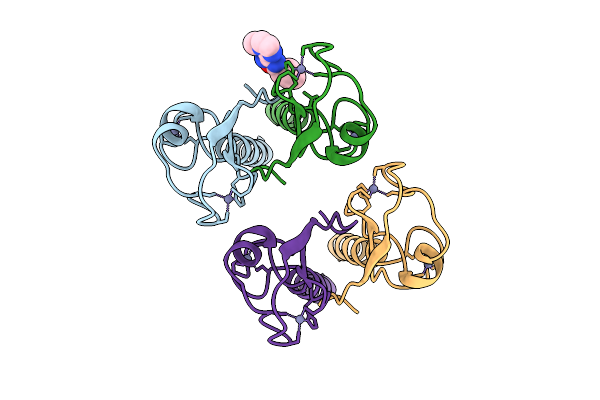 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1685106505
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, TQC |
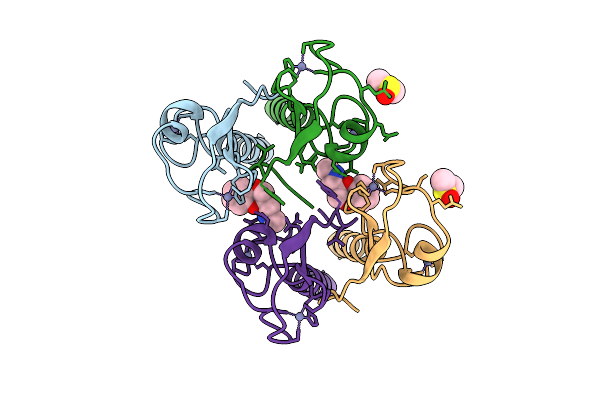 |
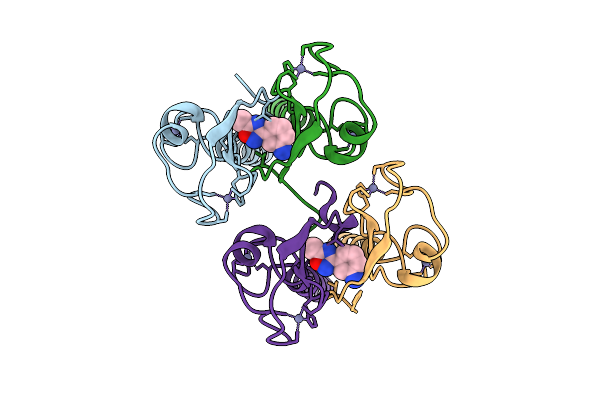
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z1891773476
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, DMS, A1BNE |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z234898049
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, A1CYF |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z2574937229
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, WJG, DMS |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z2643472210
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, JGY |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z2692078340
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, W1Y, DMS |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z275151340
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, JHP |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z275179946
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, GW1 |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z2757439080
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, Q7L |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z285233820
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, KB3, DMS |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z364577298
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, A1CYC |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z369936976
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, U0P, DMS |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z48852953
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, A1CYD, DMS |
 |
Pandda Analysis Group Deposition -- Idol Ring Domain In Complex With Z57475877
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN, S7S, DMS |

