Search Count: 59,898
 |
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Compound Zm_097
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: A1D76, DMS |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
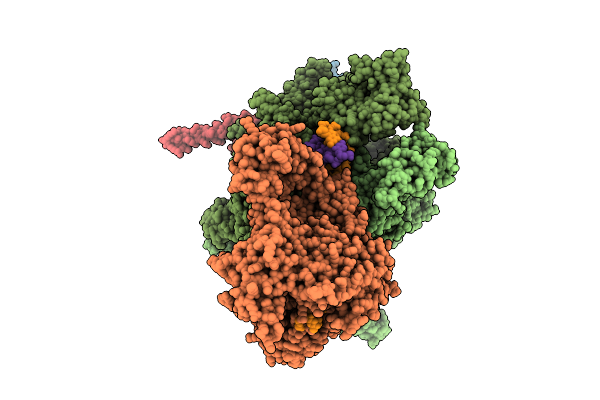
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
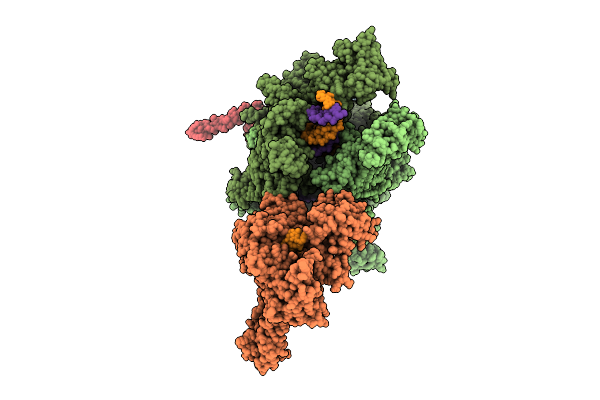
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
K115 Acetylated Human Muscle Pyruvate Kinase, Isoform M2 (Pkm2), In Complex With Fbp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: FBP, EDO, SIN, GOL, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: GOL, EDO, MG, TRS, SIN, K |
 |
K166 Acetylated Human Muscle Pyruvate Kinase, Isoform M2 (Pkm2), In Complex With Fbp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: FBP, MG |
 |
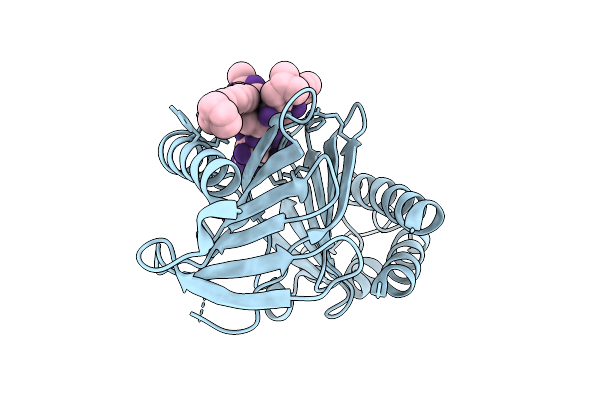
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, FMN, PUJ, GOL |
 |
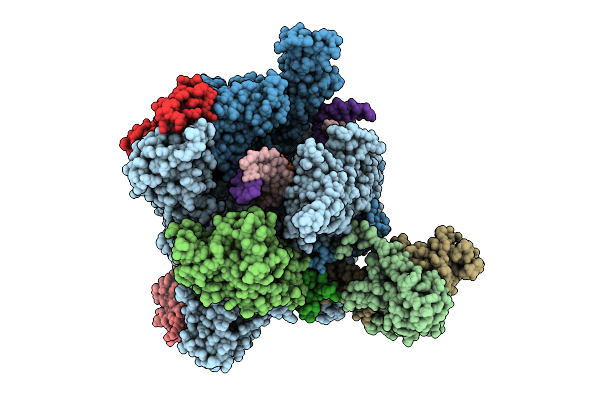
Organism: Mus musculus, Synthetic constrcut
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE |
 |
The Structure Of Rna Polymerase Ii Elongation Complex Paused At N-5 State By Actinomycin D.
Organism: Saccharomyces cerevisiae s288c, Synthetic construct, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIGASE Ligands: FES |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: ZN |
 |
Discovery Of A Bifunctional Pkmyt1-Targeting Protac Empowered By Ai-Generation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EMX, PO4, CL |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |

