Search Count: 202
 |
Organism: Rhodococcus opacus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-07-19 Classification: HYDROLASE |
 |
Organism: Beijerinckia indica subsp. indica
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: GOL |
 |
Beijerinckia Indica Beta-Fructosyltransferase Variant H395R/F473Y In Complex With Fructose
Organism: Beijerinckia indica subsp. indica nbrc 3744
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: MG, FRU, BDF |
 |
Organism: Human immunodeficiency virus type 1 bh10
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Haloarcula Marismortui Cheb With Glutathione S-Transferase Expression Tag
Organism: Schistosoma japonicum, Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2022-07-20 Classification: TRANSFERASE, HYDROLASE |
 |
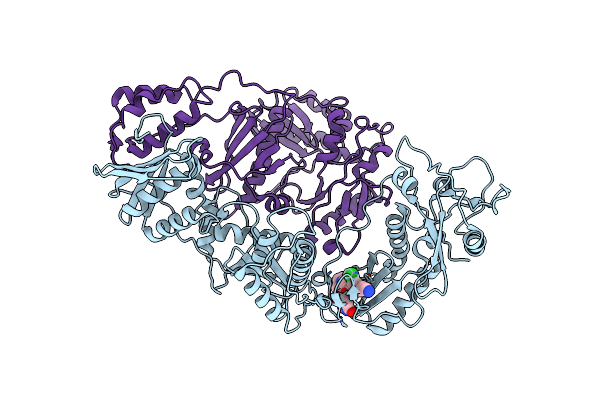
Crystal Structure Of Hiv-1 Integrase Complexed With (2S)-2-(Tert-Butoxy)-2-(5-{2-[(2-Chloro-6-M Ethylphenyl)Methyl]-1,2,3,4-Tetrahydroisoquinolin-6-Yl}-4- (4,4-Dimethylpiperidin-1-Yl)-2-Methylpyridin-3-Yl)Acetic Acid
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: O20, SO4 |
 |
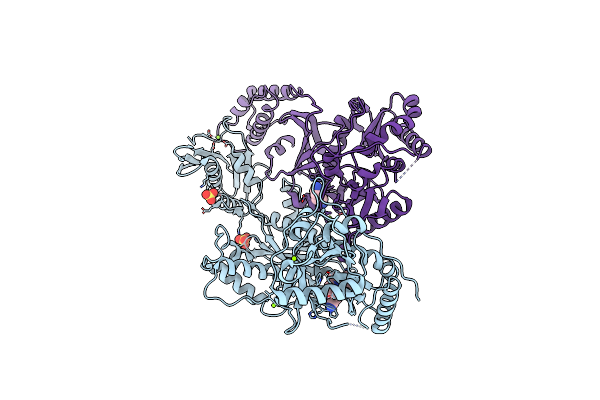
Crystal Structure Of Hiv-1 Reverse Transcriptase In Complex With (E)-3-(3-Chloro-5-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)-4-(2-Morpholinoethoxy)Phenoxy)Phenyl)Acrylonitrile (Jlj530)
Organism: Human immunodeficiency virus type 1, Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2022-03-16 Classification: HYDROLASE, Transferase/Viral Protein Ligands: 9UV |
 |
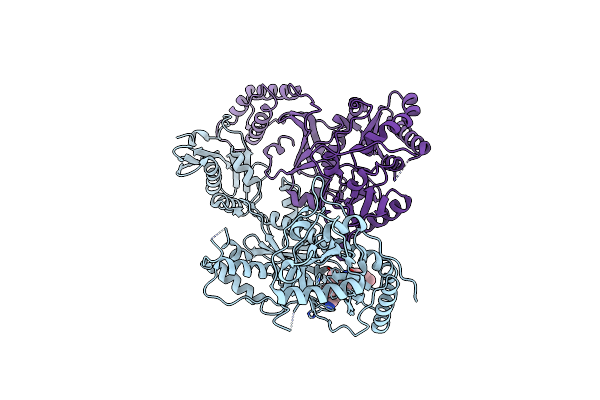
Crystal Structure Of Hiv-1 Reverse Transcriptase In Complex With 6-((2-((4-Cyanophenyl)Amino)Pyrimidin-4-Yl)Amino)-5,7-Dimethylindolizine-2-Carbonitrile (Jlj604)
Organism: Human immunodeficiency virus type 1, Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2022-03-16 Classification: HYDROLASE, Transferase/Viral Protein Ligands: 9WI, SO4, MG, PUT |
 |
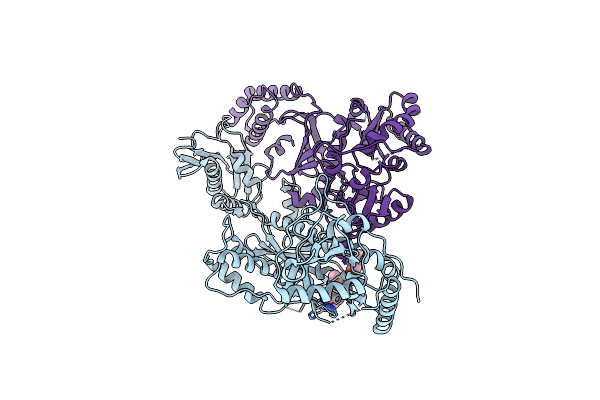
Crystal Structure Of Hiv-1 Reverse Transcriptase In Complex With (E)-4-((4-((4-(2-Cyanovinyl)-2,6-Dimethylphenyl)Amino)-6-(3-Morpholinopropoxy)-1,3,5-Triazin-2-Yl)Amino)Benzonitrile (Jlj564)
Organism: Human immunodeficiency virus type 1, Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2022-03-16 Classification: HYDROLASE, Transferase/Viral Protein Ligands: 9W3 |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase Y181C Variant In Complex With (E)-4-((4-((4-(2-Cyanovinyl)-2,6-Dimethylphenyl)Amino)-6-(3-Morpholinopropoxy)-1,3,5-Triazin-2-Yl)Amino)Benzonitrile (Jlj564)
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2022-03-16 Classification: hydrolase, transferase/viral protein Ligands: 9W3 |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase K103N/Y181C Variant In Complex With (E)-4-((4-((4-(2-Cyanovinyl)-2,6-Dimethylphenyl)Amino)-6-(3-Morpholinopropoxy)-1,3,5-Triazin-2-Yl)Amino)Benzonitrile (Jlj564)
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2022-03-16 Classification: hydrolase, transferase/viral protein Ligands: 9W3, MG |
 |
Crystal Structure Of Hiv-1 Y181C Mutant Reverse Transcriptase In Complex With 5-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)Phenoxy)-7-Fluoro-2-Naphthonitrile (Jlj635), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus type 1 group m subtype b, Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-03-16 Classification: hydrolase, transferase/viral protein Ligands: M9A, SO4 |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Sophorose
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With 1-Deoxynojirimycin
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: NOJ, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With P-Nitrophenyl-Alpha-D-Glucopyranoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: PNG, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Methyl Alpha-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: GYP, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Ethyl Alpha-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: BW3, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Phenyl Alpha-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: 7OL, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Methyl Beta-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: MGL, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With N-Octyl-Beta-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: BOG, CA |

