Search Count: 1,15,929
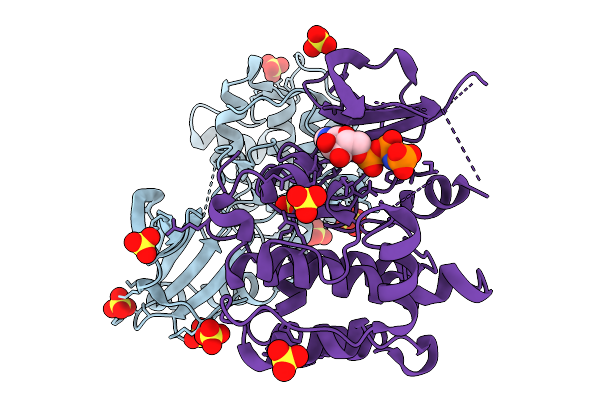 |
Crystal Structure Of Serine/Threonine-Protein Kinase (Aek1) T376D, S395D Mutant From Trypanosoma Brucei (Amp-Pnp)
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: SO4, ANP |
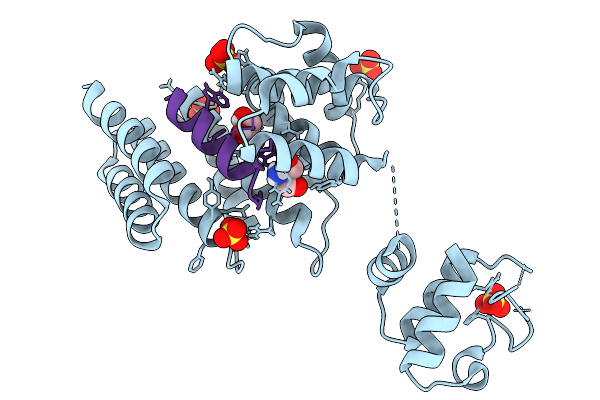 |
Crystal Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3 In Complex With Rgg3Bp13
Organism: Streptococcus thermophilus lmg 18311, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2026-02-04 Classification: DNA BINDING PROTEIN Ligands: SO4, GOL, TRS |
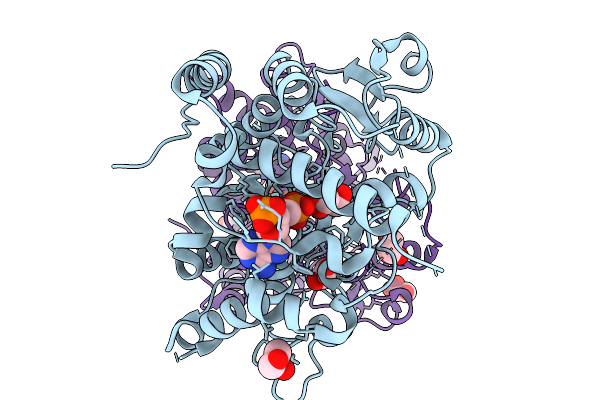 |
Crystal Structure Of Sorghum Sulfotransferase Lgs1 Reveals Sulfation-Assisted Bc-Ring Formation In Strigolactone Biosynthesis
Organism: Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: A3P, GOL |
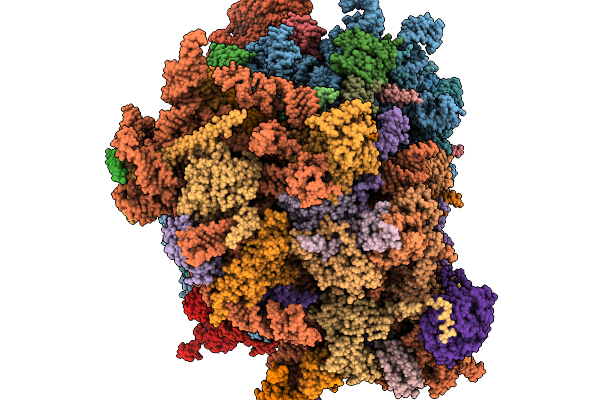 |
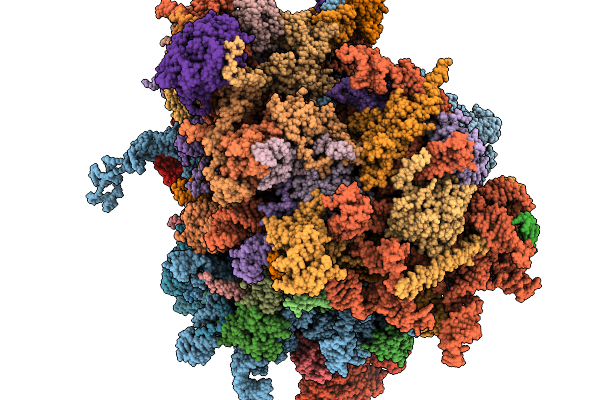
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, ZN |
 |
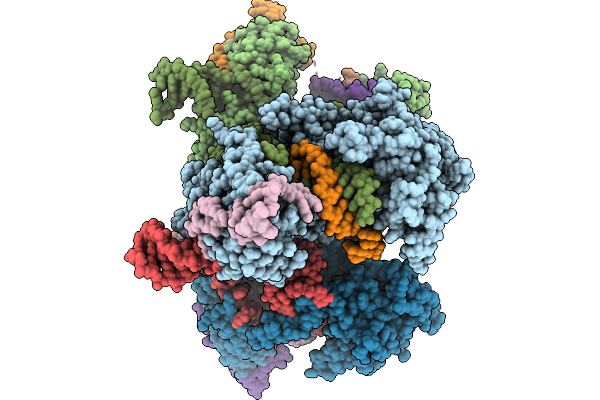
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli nctc 86
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2026-02-04 Classification: ANTIVIRAL PROTEIN |
 |
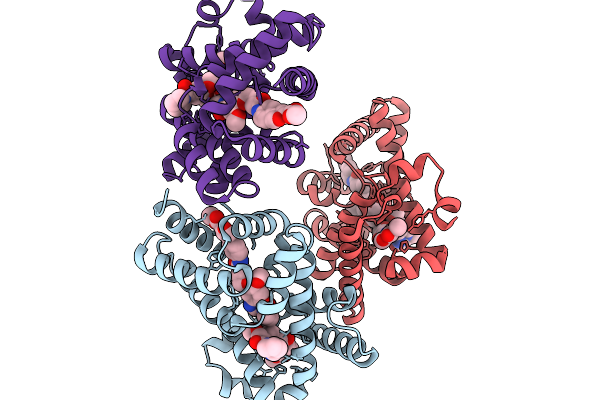
Organism: Escherichia coli nctc 86
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2026-02-04 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
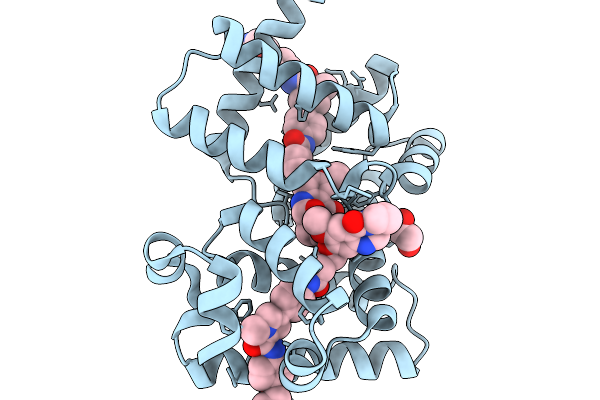
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-04-154 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2026-02-04 Classification: ANTIMICROBIAL PROTEIN Ligands: A1I1F, A1IZN |
 |
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-04-161 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2026-02-04 Classification: ANTIMICROBIAL PROTEIN Ligands: A1I0A, A1I0B, EDO |
 |
Organism: Metagenome, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Organism: Tissierellia bacterium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN Ligands: ZN, MG |
 |
Organism: Metagenome, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
Organism: Nitrospirota bacterium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Resolution:2.99 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN/DNA |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN/DNA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2026-02-04 Classification: DNA Ligands: K, SPM, PEG, PG4, PGE |
 |
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Bisphenol B
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: H3W |
 |
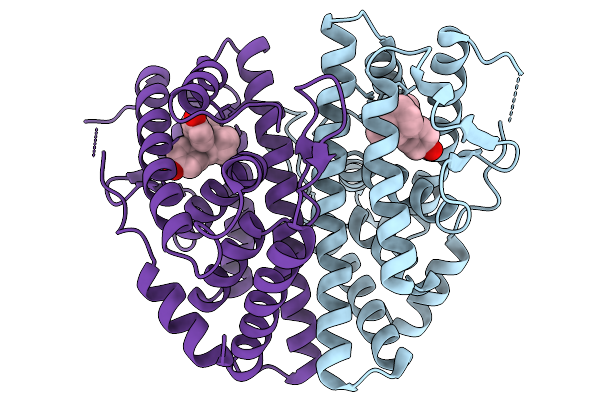
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Bisphenol Z
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: BPZ |
 |
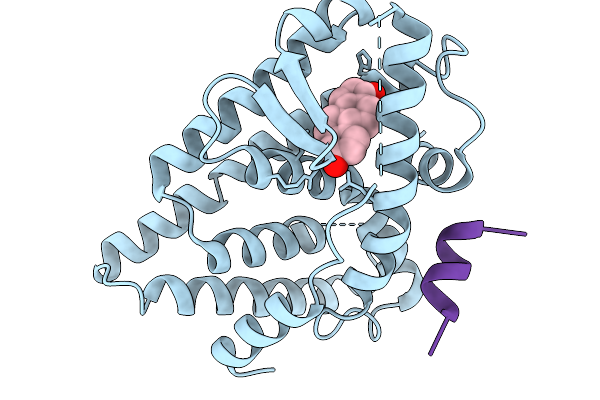
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Bisphenol Tmc
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: A1BVK, A1BWR |
 |
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Estradiol And D22 13Mer
Organism: Melanotaenia fluviatilis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: EST |

