Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
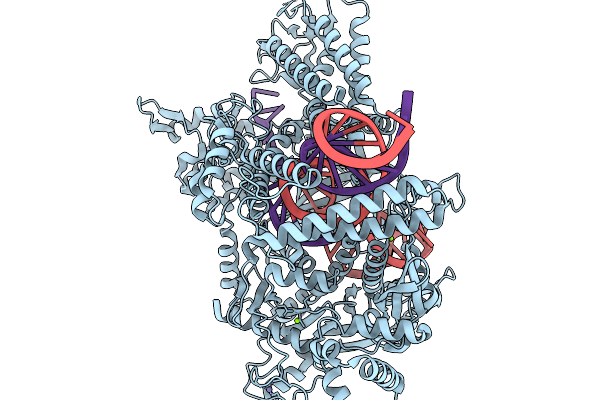
Organism: Bacteroidales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA Ligands: MG |
 |
Organism: Bacteroidales, Bacteroidales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA Ligands: MG, ZN |
 |
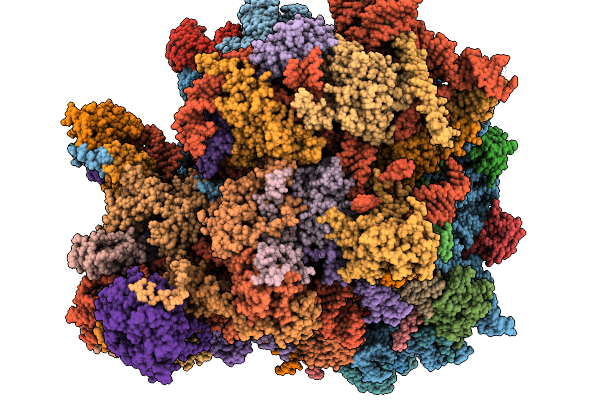
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: RNA binding protein/RNA Ligands: MG, SO4 |
 |
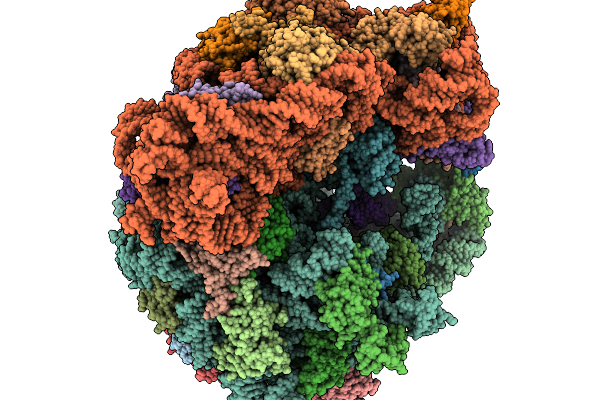
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME Ligands: GDP, MG, K, ZN, PUT, HYG, SPD, 3HE, ANM, NA |
 |
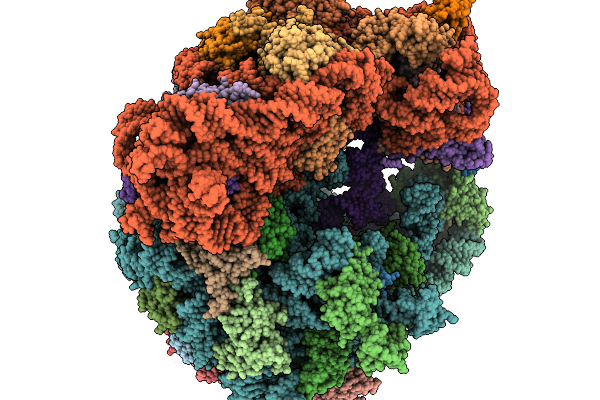
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Release Factor Bound In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
 |
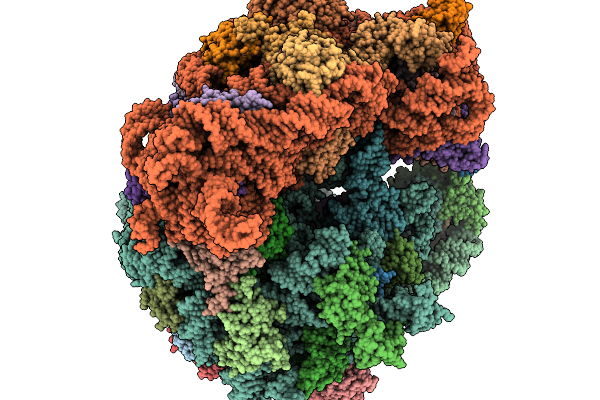
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
 |
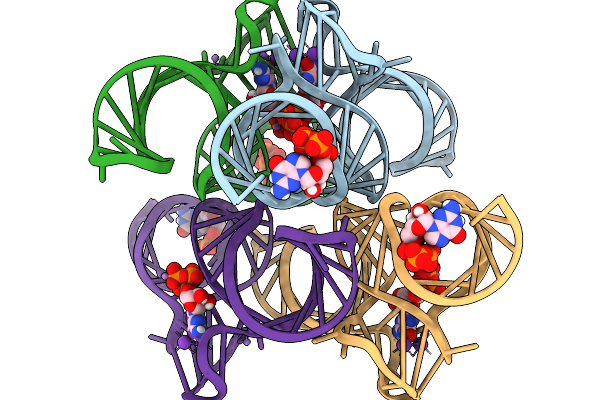
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Trna-Tyr In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
 |
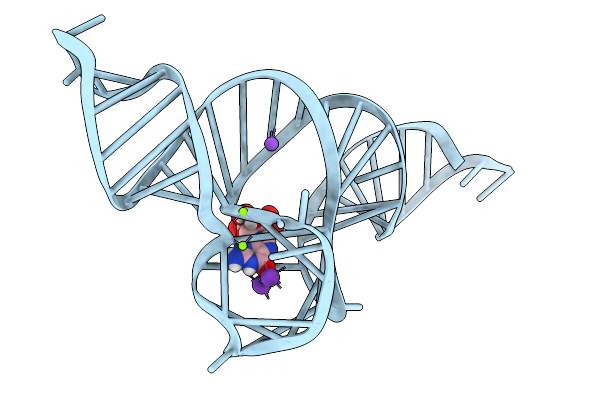
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: RNA Ligands: GTP, K, MN |
 |
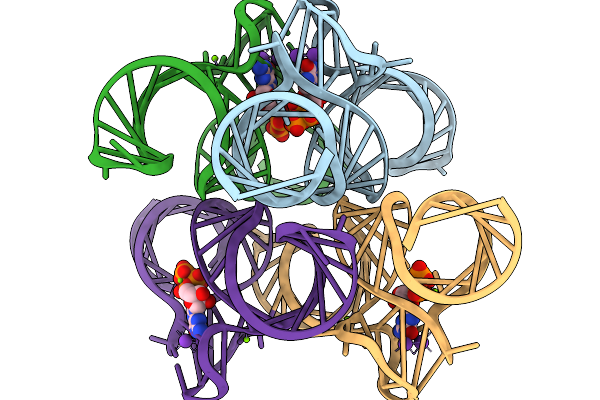
Crystal Structure Of The Class V (Uu) Gtp Aptamer Variant In Complex With Gtp
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: RNA Ligands: GTP, K, MG, NA |
 |
Crystal Structure Of The Class V (G61A) Gtp Aptamer Variant In Complex With Gtp
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: RNA Ligands: K, GTP, MG |
 |
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
 |
Pr1 Phage Heterodimeric Dna Ligase In Complex With 21-Mer Nicked Dna Containing Phage Nick Sequence
Organism: Providencia phage vb_pres_pr1
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN Ligands: NMN, AMP, ZN |
 |
Asymmetric Unit From Transcribing Double-Layered Particle Of Bacteriophage Phi6
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
 |
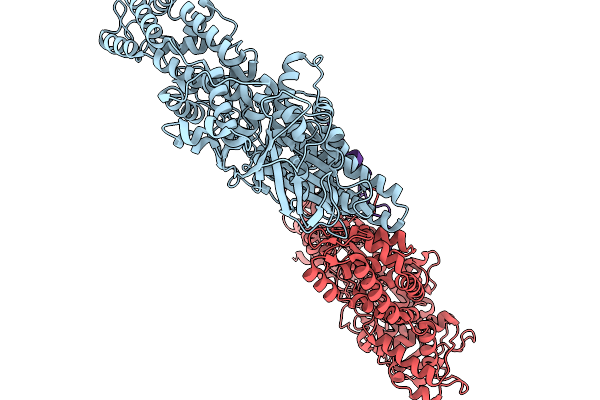
Asymmetric Unit From Transcribing Single-Layered Particle Of Bacteriophage Phi6
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
 |
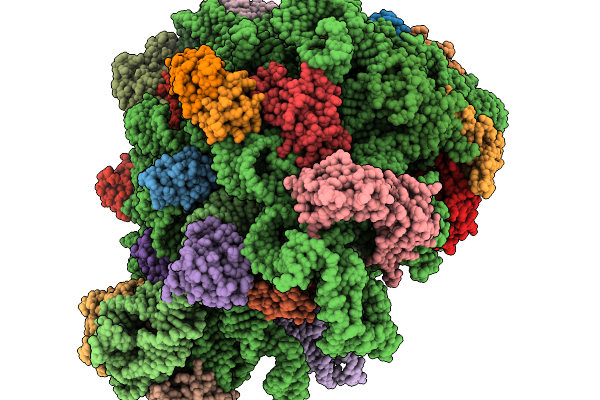
Asymmetric Unit From Transcribing Single-Layered Particle Of Bacteriophage Phi6 In An Over-Expanded State
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
 |
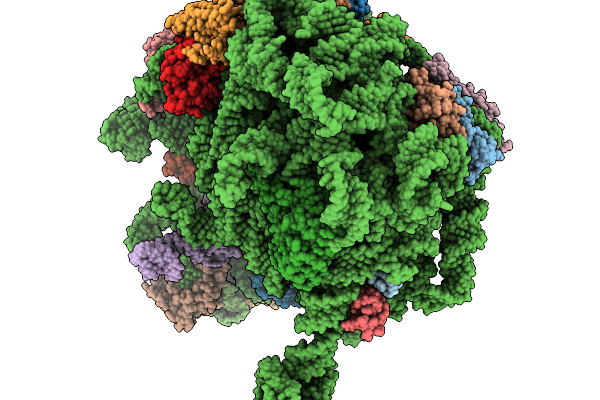
Structure Of Wt E.Coli Ribosome With Complexed Filament Nascent Chain At Length 47, With P-Site Trna
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
 |
Structure Of Wt E.Coli Ribosome With Complexed Filament Nascent Chain At Length 31, With P-Site Trnas
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: SPLICING |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: SPLICING Ligands: HOH |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: SPLICING |