Search Count: 304
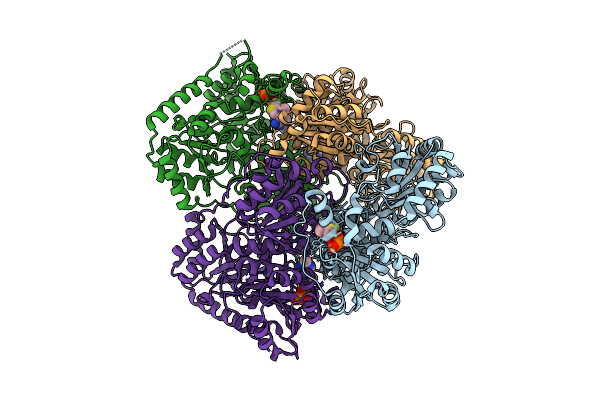 |
Ancestrally Reconstructed Acetolactate Synthase - Ancestor N259 With Bound Thdp And Magnesium.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: TPP, MG |
 |
A Thdp-Dependent Enzyme Belonging To The 1-Deoxy-D-Xylulose-5-Phosphate Synthase (Dxps)-Like Subfamily
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, TPP |
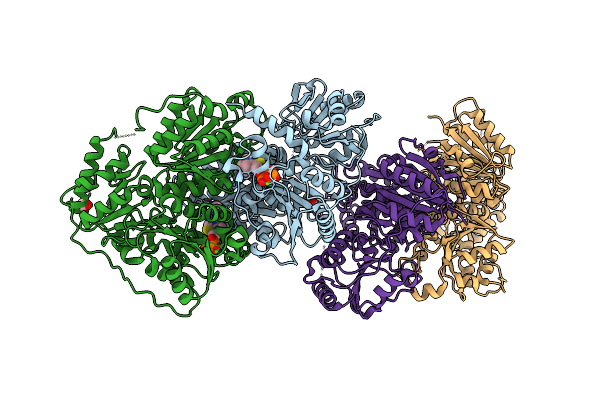 |
Alpha-Ketoisovalerate Decarboxylase (Kivd) From Synechocystis Sp. Pcc 6803 With Substitution S286T
Organism: Lactococcus lactis subsp. lactis
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: LYASE Ligands: TPP, EDO, MG |
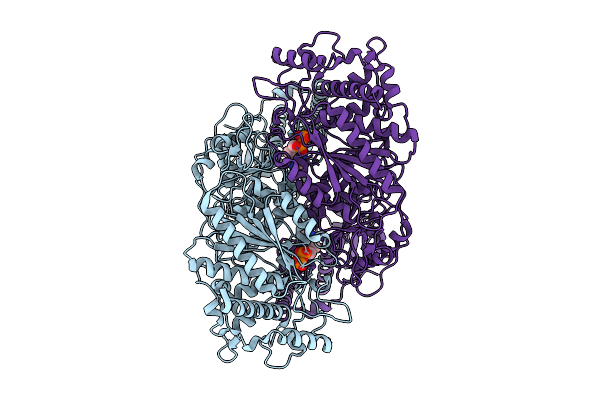 |
Cryo-Em Structure Of Candidatus Saccharibacterium Phosphoketolase Complexed With Thiamine Diphosphate
Organism: Candidatus saccharibacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: TPP |
 |
Organism: Listeria monocytogenes 10403s
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: TPP, MG |
 |
Cryo-Em Structure Of The Helicobacter Pylori Oordabc Complex In The Apo-Form
Organism: Helicobacter pylori
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: SF4, MG, TPP |
 |
Organism: Helicobacter pylori
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: SF4, A1D65, MG, TPP |
 |
Crystal Structure Of The Decarboxylase Kdc4427 From Enterobacter Sp. Cgmcc 5087
Organism: Enterobacter sp. cgmcc 5087
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-05-21 Classification: LYASE Ligands: TPP, MG |
 |
Crystal Structure Of The Decarboxylase Kdc4427 Mutant E468L From Enterobacter Sp. Cgmcc 5087
Organism: Enterobacter sp. cgmcc 5087
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-05-21 Classification: LYASE Ligands: TPP, MG |
 |
Crystal Structure Of The Decarboxylase Kdc4427 Mutant E468L In Complex With Indole-3-Pyruvic Acid
Organism: Enterobacter sp. cgmcc 5087
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-05-21 Classification: LYASE Ligands: TPP, MG, 3IO |
 |
Crystal Structure Of The Decarboxylase Kdc4427 Mutant E468L In Complex With Phenylpyruvic Acid
Organism: Enterobacter sp. cgmcc 5087
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2025-05-21 Classification: LYASE Ligands: TPP, MG, PPY |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, CL, TPP, DNA, MG |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, TPP, DNA, MG, CL |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, CL, TPP, DNA, EDO, MG |
 |
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: TPP |
 |
Crystal Structure Of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
Organism: Herbiconiux sp. salv-r1
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2025-02-12 Classification: LYASE Ligands: TPP, MG |
 |
Crystal Structure Of A Benzaldehyde Lyase Mutant M3 From Herbiconiux Sp. Salv-R1
Organism: Herbiconiux sp. salv-r1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-02-12 Classification: LYASE Ligands: PEG, MES, TPP, MG |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2025-01-01 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, A1LX0, MG, TPP |
 |
Crystal Structure Of Phosphonopyruvate Decarboxylase Rhief From Bacillus Subtilis Atcc6633 In Complex With Thiamine Pyrophosphate
Organism: Bacillus spizizenii atcc 6633 = jcm 2499
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-12-18 Classification: LYASE Ligands: MG, TPP, SPV |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, MG |

