Search Count: 5,297
 |
Organism: Homo sapiens, Bungarus multicinctus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TOXIN Ligands: EDO |
 |
Organism: Homo sapiens, Naja kaouthia
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TOXIN Ligands: GOL, CL, K, NA, SO4 |
 |
Organism: Homo sapiens, Naja kaouthia
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TOXIN Ligands: CL, GOL, TRS, SO4, NA |
 |
Cryo-Em Structure Of The Type Ii Secretion System Protein From Acidithiobacillus Caldus
Organism: Acidithiobacillus caldus (strain sm-1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TOXIN |
 |
High Resolution Structure Of Cytidine Deaminase T6S Toxin From Pseudomonas Syringae
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TOXIN Ligands: ZN, PO4, EDO, NA |
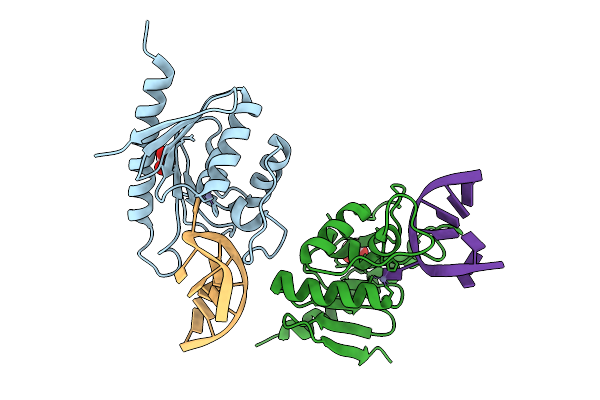 |
Cytidine Deaminase T6S Toxin From Pseudomonas Syringae Complexed With Substrate Dna
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TOXIN/DNA Ligands: ZN, EDO |
 |
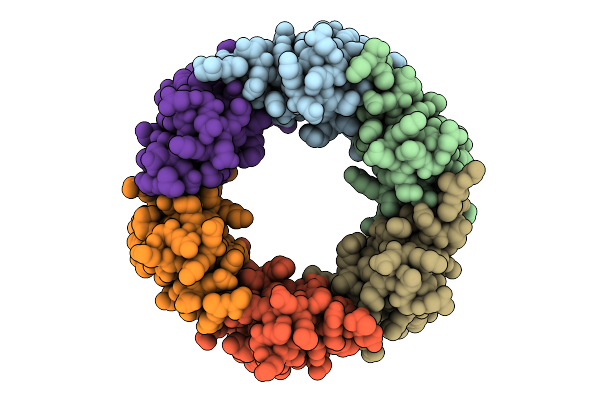
Native Structure Of The Full-Length Pesticidal Protein Cry1Ca18 At Ph7, From Crystals Formed In Vivo
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TOXIN Ligands: ZN, CA |
 |
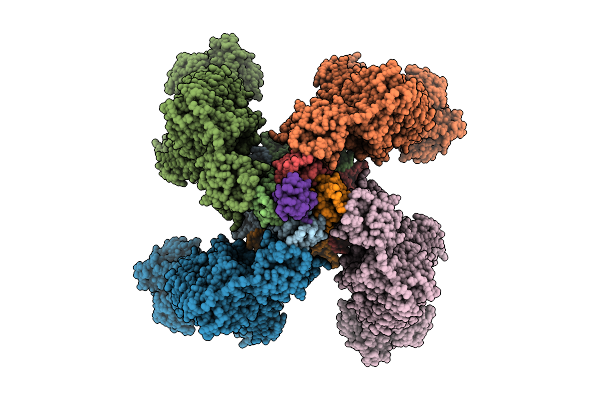 |
Organism: Homo sapiens, Mesocricetus auratus, Scolopendra polymorpha
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: ATP, GBM |
 |
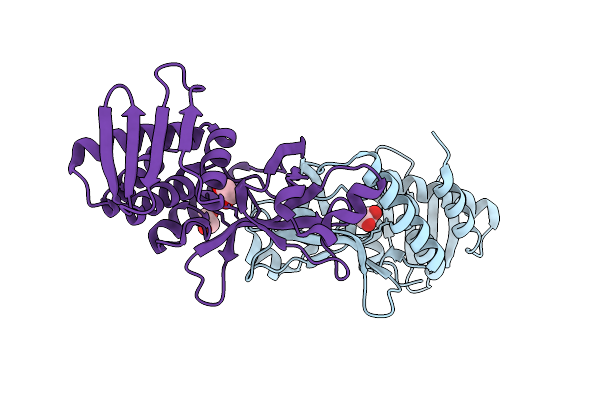
Cryoem Structure Of De Novo Antibody Fragment Scfv 6 With C. Difficile Toxin B (Tcdb)
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN |
 |
Structure Of The Tela-Associated Type Vii Secretion System Chaperone Sir_0168
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: EDO, CL |
 |
Organism: Streptococcus intermedius
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: FMT, ACY |
 |
Structure Of The Telb-Associated Type Vii Secretion System Chaperone Sir_0178
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: CL |
 |
Structure Of The Tela-Associated Type Vii Secretion System Chaperone Lcpa In Complex With The Chaperone Binding Site Of Tela
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: CIT |
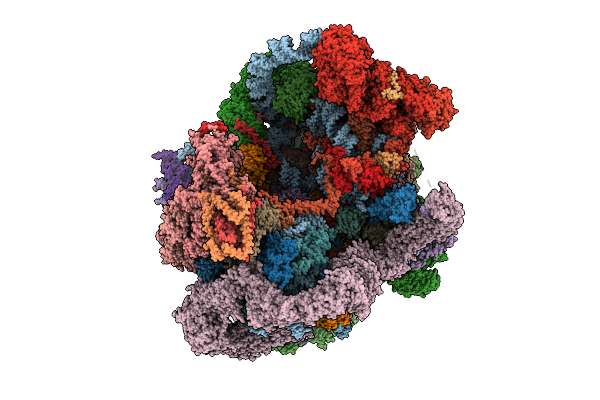 |
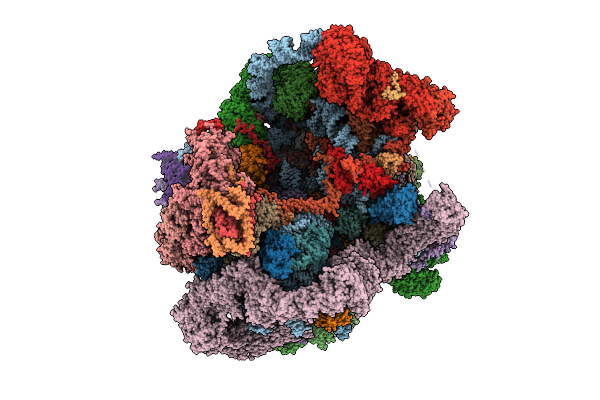
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
 |
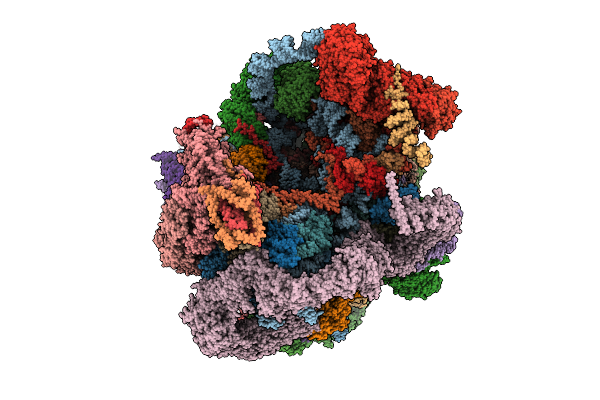
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
 |
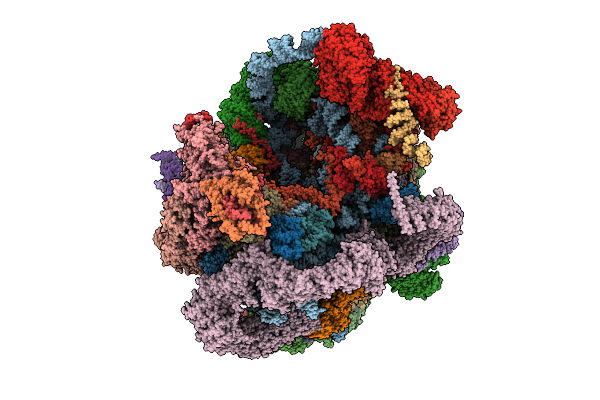
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
 |
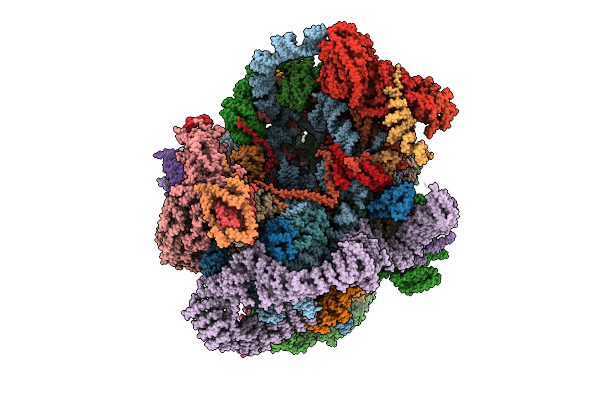
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |
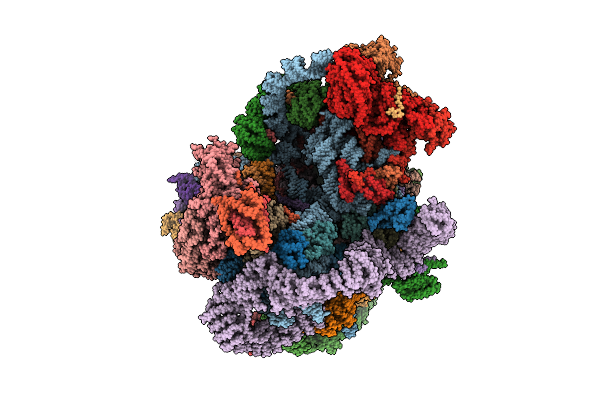 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |

