Search Count: 42,226
 |
Structure Of E.Coli Ribosome With Nascent Chain At Linker Length Of 31 Amino Acids, With Mrna, P-Site And A-Site Trnas
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME |
 |
Crystal Structure Of Dasr In Complex With A Synthetic Dasr-Binding Rna Aptamer
Organism: Streptomyces coelicolor, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: BR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME Ligands: K, MG, NA, SPD, PUT, ZN, HYG |
 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2026-01-28 Classification: SIGNALING PROTEIN Ligands: A1IZQ, DMS |
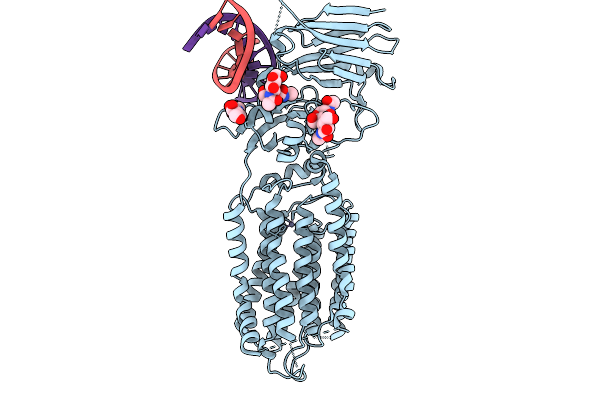 |
Organism: Caenorhabditis elegans, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.99 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/RNA Ligands: NAG, ZN |
 |
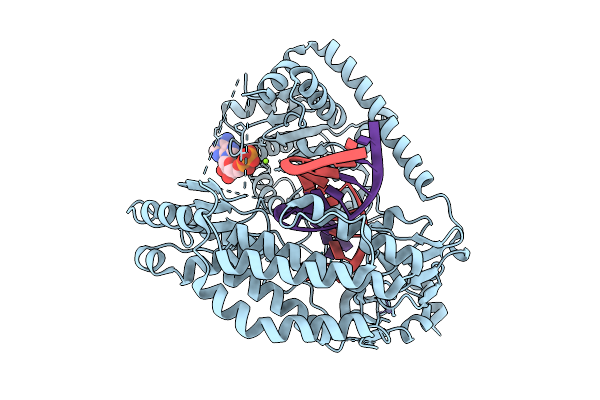
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: A1EY1, ECC |
 |
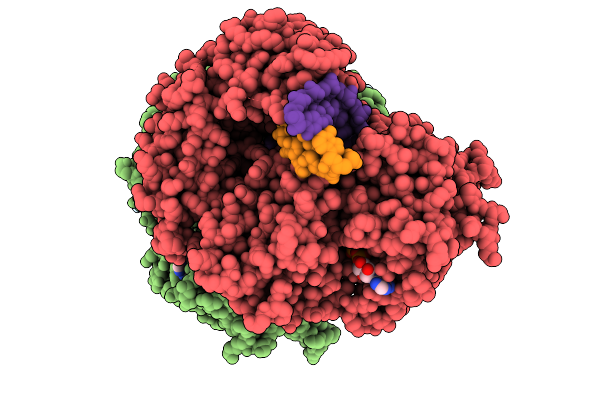
Organism: Homo sapiens, Uncultured archaeon
Method: ELECTRON MICROSCOPY Resolution:3.36 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM/RNA Ligands: ADP, ZN, MG |
 |
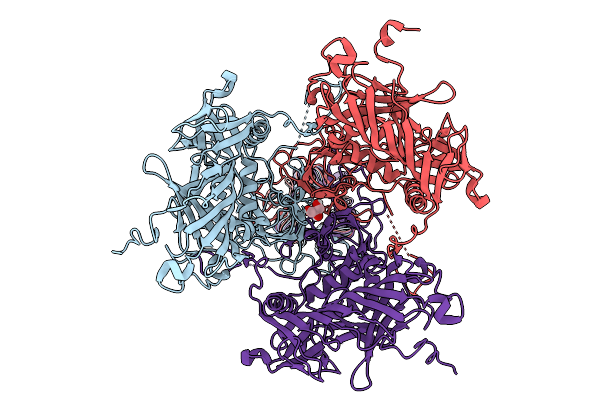
Organism: Homo sapiens, Uncultured archaeon
Method: ELECTRON MICROSCOPY Resolution:3.07 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM/RNA Ligands: ADP, ZN, MG |
 |
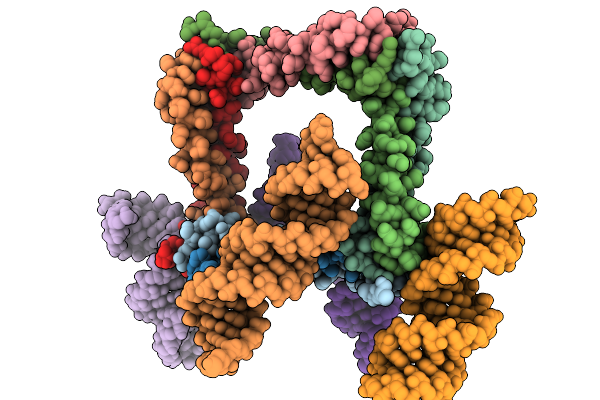
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2026-01-28 Classification: PROTEIN TRANSPORT Ligands: SO4, GOL |
 |
Organism: Hepatitis delta virus, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2026-01-28 Classification: VIRUS |
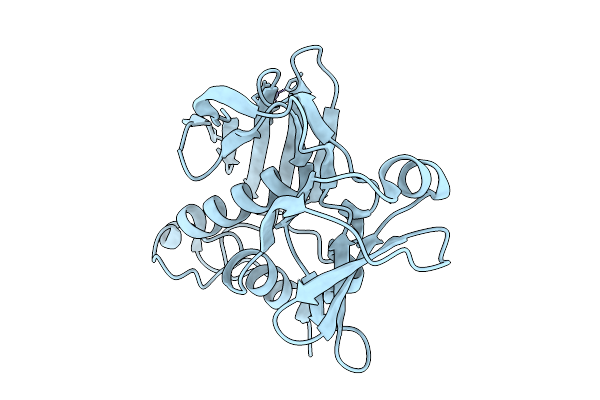 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2026-01-28 Classification: TOXIN Ligands: ZN |
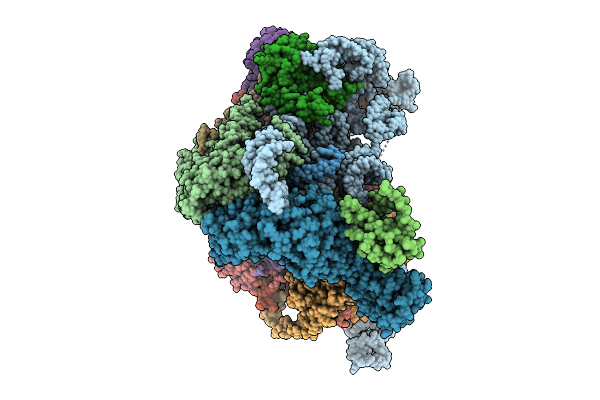 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME |
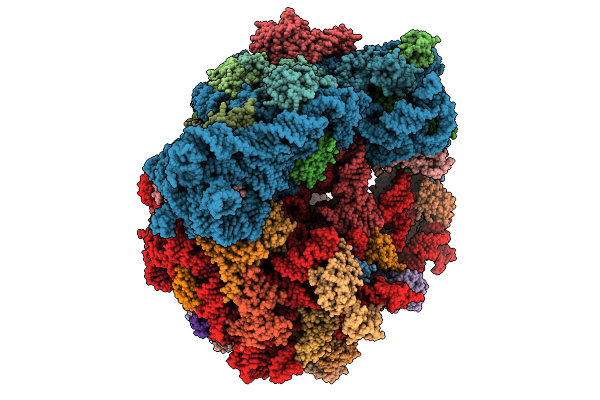 |
Organism: Escherichia coli, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME Ligands: ZN, K, MG, PRO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2026-01-28 Classification: TRANSCRIPTION Ligands: A1JIF, PEG, ZN, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: GOL, PEG, A1JI1, ZN, SO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: GDP |
 |
Cryo-Em Structure Of The Catalytic Core Of Human Telomerase At The Initiation State Of The Repeat Addition Cycle
Method: ELECTRON MICROSCOPY
Resolution:3.53 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: 1GC |
 |
Cryo-Em Structure Of The Catalytic Core Of Human Telomerase At The Elongation State Of The Repeat Addition Cycle
Method: ELECTRON MICROSCOPY
Resolution:3.20 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: A1ACD |
 |
Cryo-Em Structure Of The Catalytic Core Of Human Telomerase At The Pre-Termination State Of The Repeat Addition Cycle
Method: ELECTRON MICROSCOPY
Resolution:3.80 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: 1GC |
 |
Organism: Homo sapiens, Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Resolution:2.10 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN |

