Search Count: 18,720
 |
Organism: Homo sapiens, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2026-01-28 Classification: SIGNALING PROTEIN Ligands: A1IZQ, DMS |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2026-01-28 Classification: PROTEIN TRANSPORT Ligands: SO4, GOL |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: TRANSPORT PROTEIN/Isomerase Ligands: CFF, CA, 117, ZN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2026-01-28 Classification: TOXIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2026-01-28 Classification: TRANSCRIPTION Ligands: A1JIF, PEG, ZN, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: GOL, PEG, A1JI1, ZN, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: DQV, NCA |
 |
Organism: Escherichia phage lambda, Escherichia coli o157:h7 str. edl933
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2026-01-28 Classification: ISOMERASE Ligands: A1E1M, GOL, CL, MES |
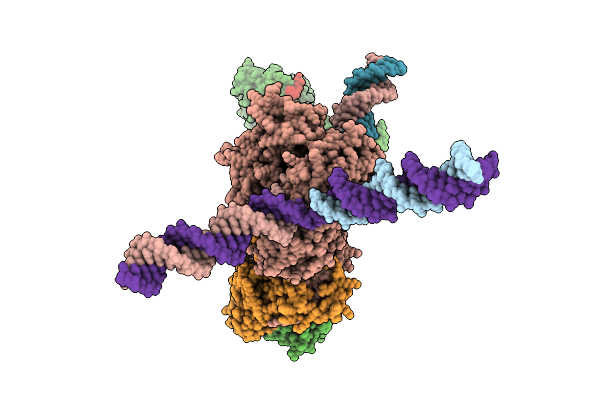 |
Cryo-Em Structure Of F-Box Helicase 1 (Fbh1) Bound To An Scf Ubiquitin Ligase Complex And A 3-Way Dna Fork (Consensus Structure)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: ISOMERASE/DNA Ligands: AGS, MG, ZN |
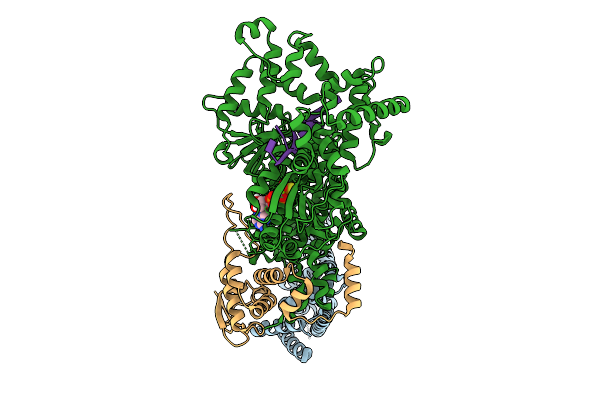 |
Cryo-Em Structure Of F-Box Helicase 1 (Fbh1) Bound To An Scf Ubiquitin Ligase Complex And A 3-Way Dna Fork (Body Structure)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: ISOMERASE/DNA Ligands: AGS, MG, ZN |
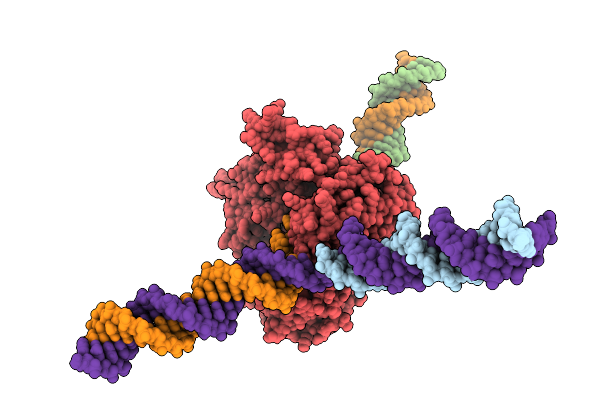 |
Cryo-Em Structure Of F-Box Helicase 1 (Fbh1) Bound To An Scf Ubiquitin Ligase Complex And A 3-Way Dna Fork (Substrate Structure)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: ISOMERASE/DNA Ligands: AGS, MG, ZN |
 |
Organism: Brugia malayi
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2026-01-21 Classification: TRANSFERASE Ligands: SO4, CL |
 |
Organism: Homo sapiens, Micrurus tener tener
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Micrurus tener tener
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN |
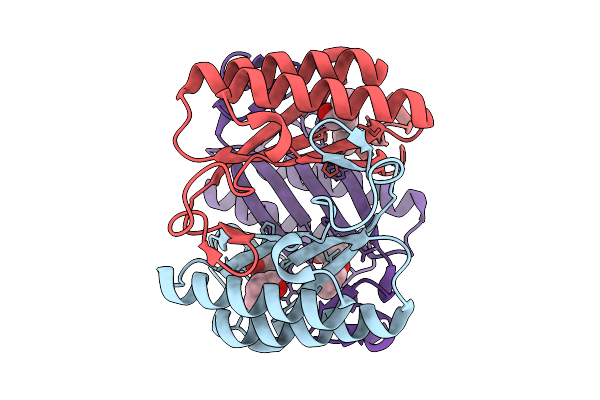 |
X-Ray Crystallographic Structure Of 4-Ot (F11) In Complex With 2-Hydroxycinnamaldehyde
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2026-01-21 Classification: LYASE Ligands: A1IZH |
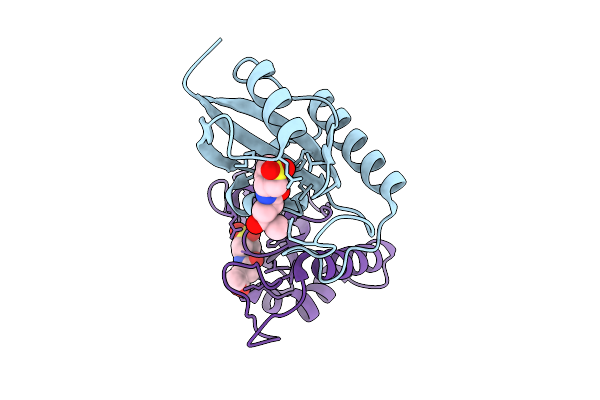 |
Human Pin1 (Peptidyl-Prolyl Cis-Trans Isomerase) Catalytic Domain Complexed With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9T |
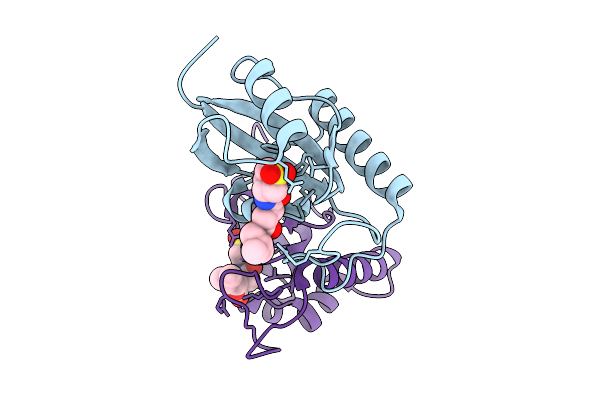 |
Human Pin1 (Peptidyl-Prolyl Cis-Trans Isomerase) Catalytic Domain In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9U |
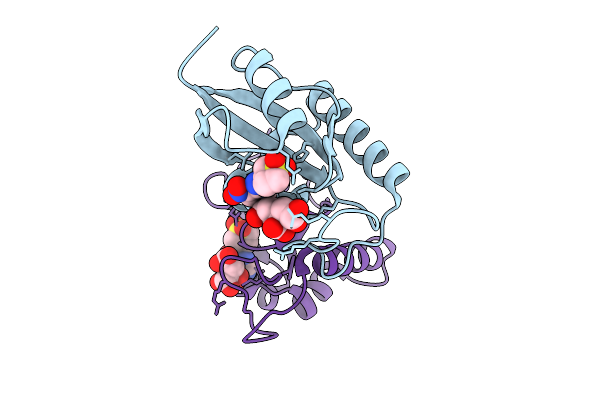 |
Crystal Structure Of Human Pin1 Catalytic Domain In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9W, CIT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: UHB |

