Search Count: 5,725
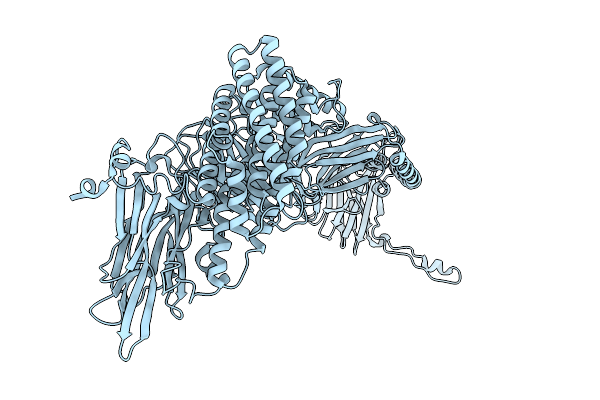 |
Native Structure Of The Full-Length Pesticidal Protein Cry8Ba2, From Crystals Formed In Vivo (Form 1)
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TOXIN |
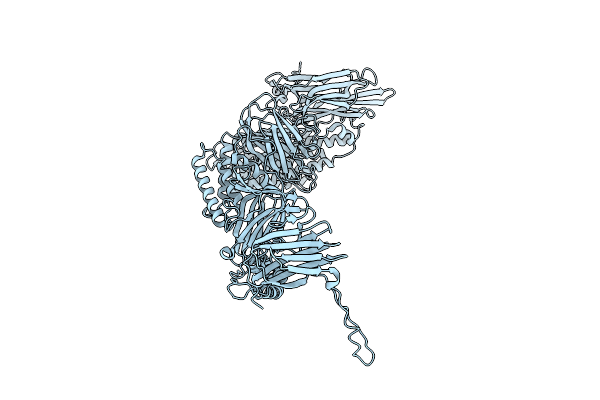 |
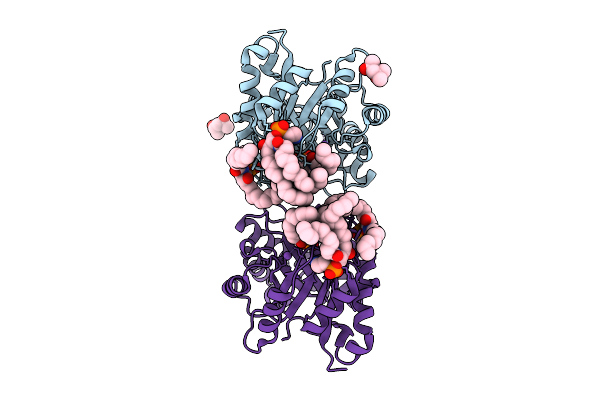
Native Structure Of The Full-Length Pesticidal Protein Cry8Ba2, From Crystals Formed In Vivo (Form 2)
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TOXIN |
 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 2.2 A Resolution From A 2-Day Old Crystal
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Substrate Sphingolipids At 2.60 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, NA, MG, MPD |
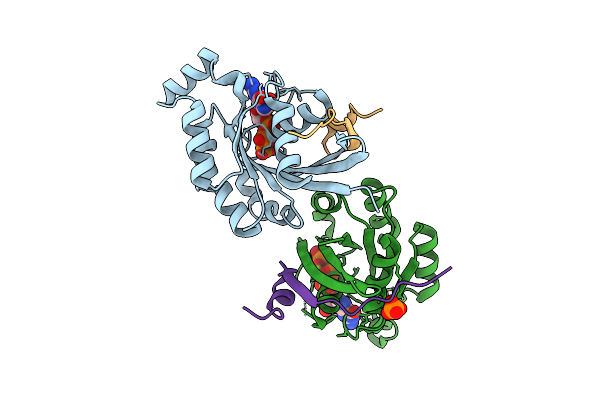 |
Crystal Structure Of Human Rac1 In Complex With The Scaffold Protein Posh (Residues 321-348)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: GNP, MG, PO4, CL |
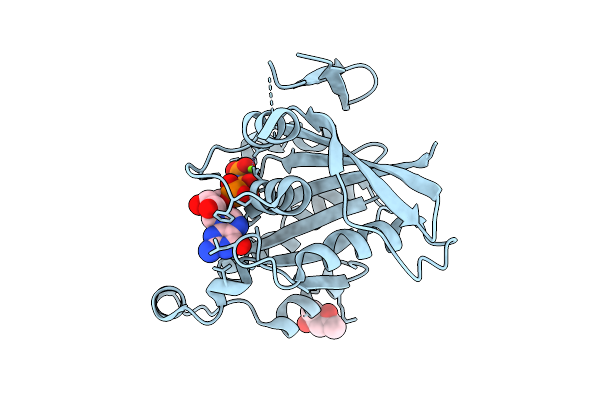 |
Crystal Structure Of Human Rac1 Fused With The Scaffold Protein Posh (Residues 319-371)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: GNP, MG, MPD |
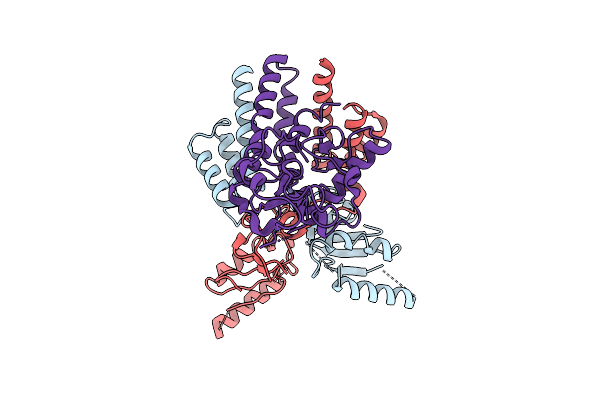 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN Ligands: ZN |
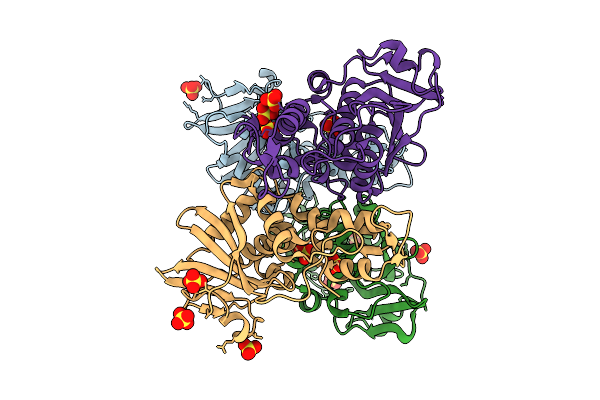 |
Organism: Mirabilis jalapa
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ANTIVIRAL PROTEIN Ligands: SO4 |
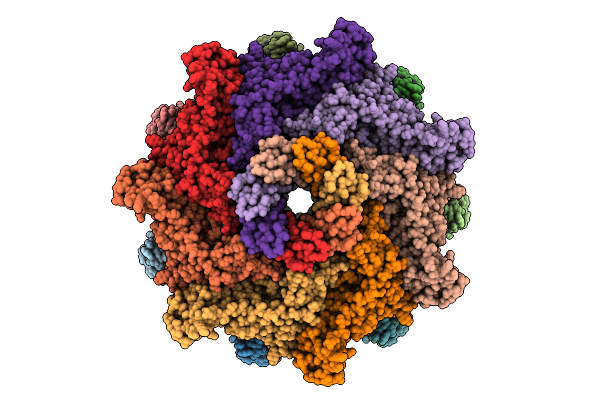 |
Organism: Aeromonas hydrophila
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
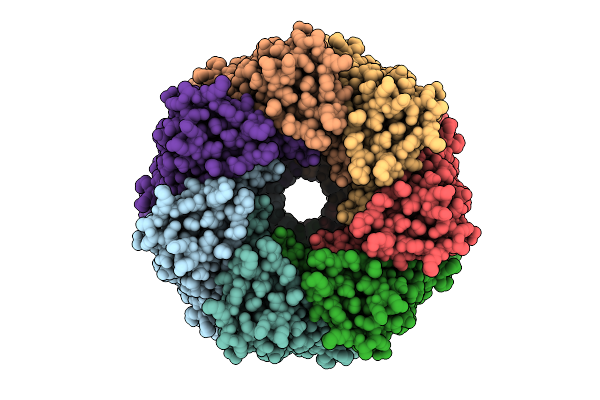 |
Cryo-Em Structure Of Pre-Pore State Iv Of Heptameric Alpha-Hemolysin In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
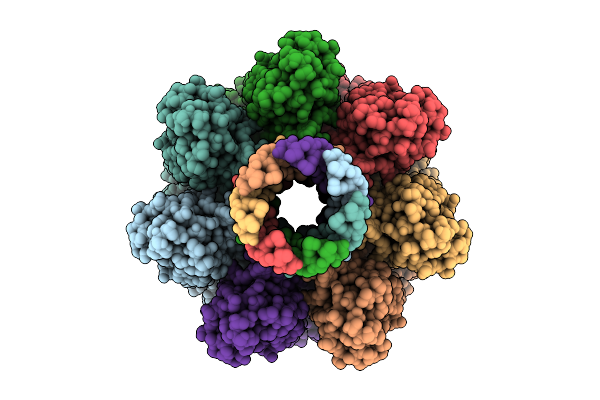 |
Cryo-Em Structure Of Alpha-Hemolysin Heptameric Pore State In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
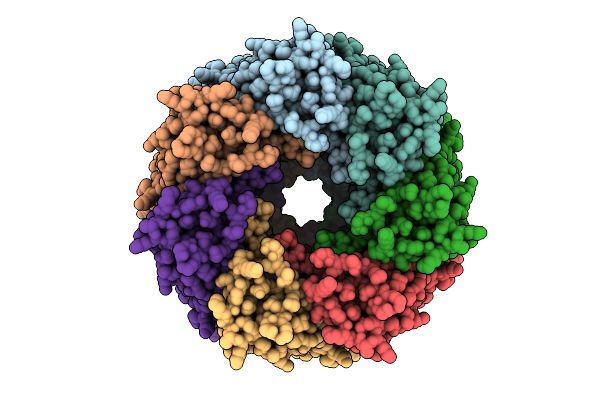 |
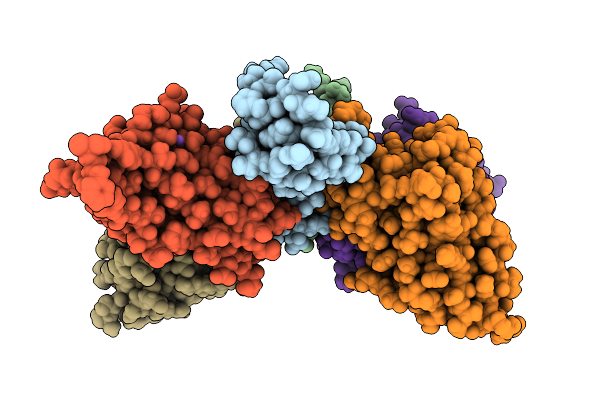
Cryo-Em Structure Of Alpha-Hemolysin Heptameric Pre-Pore State Iii In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
 |
Phosphatidylcholine Head Group Bound Alpha-Hemolysin Heptameric Pore Structure In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN Ligands: PC |
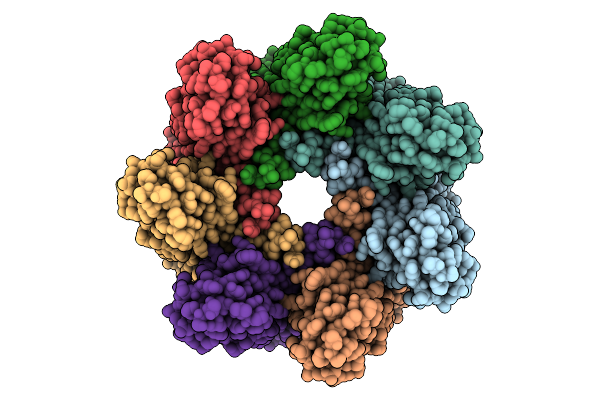 |
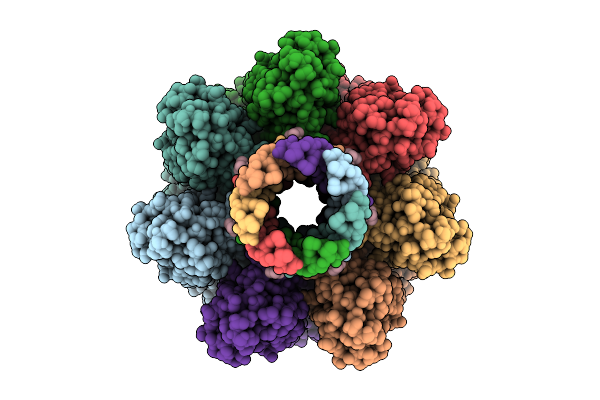
Cryo-Em Structure Of Pre-Pore State Ii Of Heptameric Alpha-Hemolysin In The Presence Of Rbcs
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
 |
Organism: Homo sapiens, Bungarus multicinctus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TOXIN Ligands: EDO |
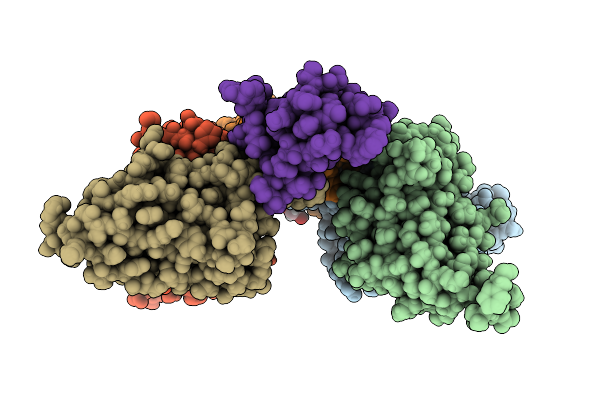
 |
Organism: Homo sapiens, Naja kaouthia
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TOXIN Ligands: GOL, CL, K, NA, SO4 |
 |
Organism: Homo sapiens, Naja kaouthia
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TOXIN Ligands: CL, GOL, TRS, SO4, NA |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: ANTITOXIN/DNA/RNA |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: ANTITOXIN/DNA/RNA Ligands: ATP |
 |
Cryo-Em Structure Of The Type Ii Secretion System Protein From Acidithiobacillus Caldus
Organism: Acidithiobacillus caldus (strain sm-1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TOXIN |

