Search Count: 82
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH4, CL |
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH3, IMD, EPE, NI, CL |
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: A1BH2, EDO |
 |
Organism: Escherichia coli o104:h4, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-05-29 Classification: TOXIN, Hydrolase Ligands: 2PE, EDO, PO4, CL |
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2024-05-01 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: CL, U4T, EDO, 2PE |
 |
Organism: Ricinus communis
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-05-01 Classification: TOXIN,HYDROLASE/INHIBITOR Ligands: ZXJ, CL, 2PE |
 |
Organism: Allochromatium vinosum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-13 Classification: HYDROLASE |
 |
Organism: Allochromatium vinosum, Synthetic construct, Streptococcus thermophilus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-13 Classification: RIBOSOME Ligands: ZN, MG |
 |
Organism: Trichlorobacter lovleyi
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-12 Classification: HYDROLASE Ligands: EDO, CL |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: PEG, GOL, IOD, CA |
 |
Crystal Structure Of Full-Length Phospholipase D From Pseudomonas Aeruginosa Pao1
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-17 Classification: HYDROLASE |
 |
Crystal Structure Of Phospholipase D From Pseudomonas Aeruginosa Pao1 Using In Situ Proteolysis
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-08-17 Classification: HYDROLASE Ligands: CA |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: HYDROLASE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: HYDROLASE |
 |
Organism: Bothrops atrox
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-12 Classification: TOXIN, HYDROLASE Ligands: DAO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: HYDROLASE |
 |
Structure Of Topi1 Inhibitor From Tityus Obscurus Scorpion Venom In Complex With Trypsin
Organism: Bos taurus, Tityus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2020-07-01 Classification: hydrolase/hydrolase inhibitor Ligands: CA, SO4 |
 |
The Tle Hydrolase Bound To The Ttr Domain Of The Vgrg Spike Of The Type 6 Secretion System
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: HYDROLASE |
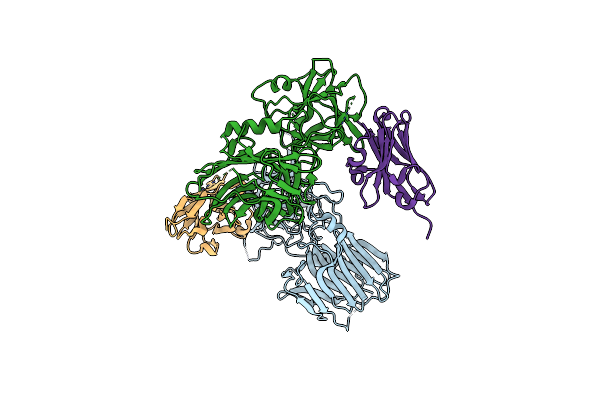 |
Crystal Structure Of Bont/B Receptor-Binding Domain In Complex With Vhh Jli-H11
Organism: Clostridium botulinum, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2020-03-04 Classification: HYDROLASE/ANTITOXIN |
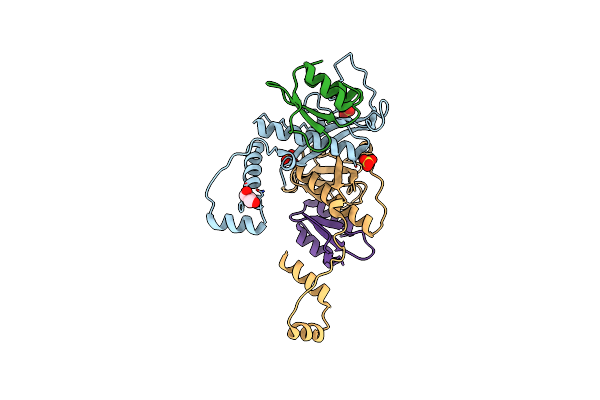 |
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-05-23 Classification: ANTITOXIN/HYDROLASE Ligands: SO4, GOL |

