Search Count: 1,009
All
Selected
 |
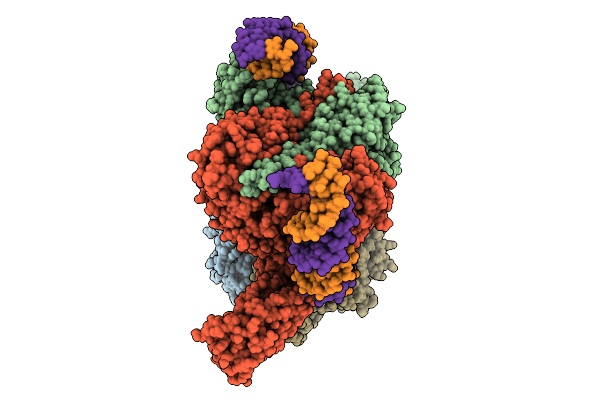
Cryoem Structure Of M. Mazei Topoisomerase Vi(A-E342Q)-Minicircle Dna Complex In Cleavage State
Organism: Methanosarcina mazei go1, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2026-02-25 Classification: ISOMERASE/DNA Ligands: CA, ANP, K |
 |
Cryoem Structure Of M. Mazei Topoisomerase Vi(A-E342Q)-Minicircle Dna Complex In Asymmetric State
Organism: Methanosarcina mazei go1, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2026-02-25 Classification: ISOMERASE/DNA Ligands: ANP, CA, K |
 |
Organism: Methanosarcina mazei go1, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2026-02-25 Classification: ISOMERASE/DNA Ligands: ANP, MG, K |
 |
Cryoem Structure Of M. Mazei Topoisomerase Vi-Minicircle Dna Complex In Partially Unfolded Transducer State
Organism: Methanosarcina mazei go1, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2026-02-25 Classification: ISOMERASE/DNA Ligands: MG, ANP, K |
 |
Cryoem Structure Of M. Mazei Topoisomerase Vi-Minicircle Dna Complex In Asymmetric State
Organism: Methanosarcina mazei go1, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2026-02-25 Classification: ISOMERASE/DNA Ligands: ANP, MG, K |
 |
Solution Nmr Structures Of Htert Dna G-Quadruplexes (G4S) In Complex With Small Molecule Cpt-11
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2026-02-11 Classification: DNA Ligands: K, CP0 |
 |
Solution Nmr Structures Of Htert Dna G-Quadruplexes (G4S) In Complex With Small Molecule Zbh-01
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2026-02-11 Classification: DNA Ligands: K, A1EQ9 |
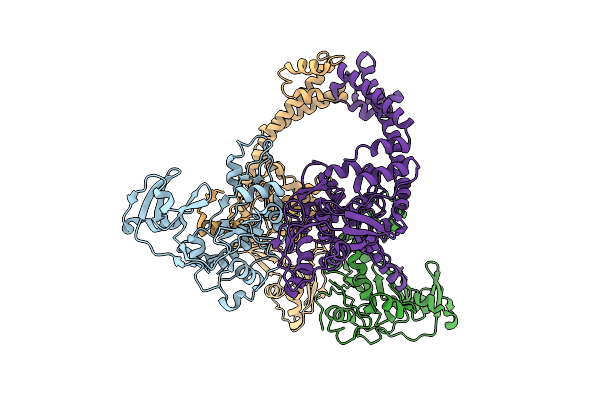 |
Organism: Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: ISOMERASE |
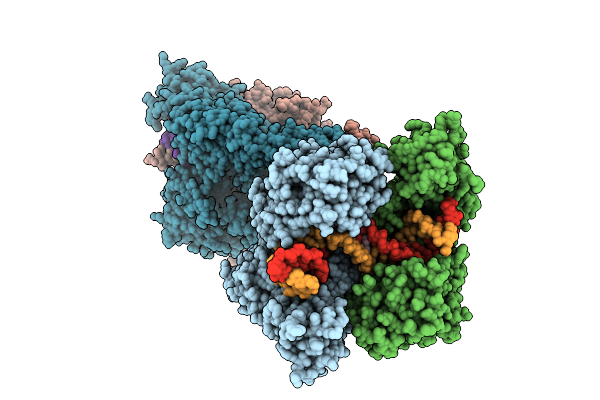 |
Crystal Structure Of Topoisomerase Iv From Klebsiella Pneumoniae In Complex With Dna And Bwc0977, A Dual-Targeting Broad-Spectrum Novel Bacterial Topoisomerase Inhibitor.
Organism: Klebsiella pneumoniae subsp. pneumoniae mgh 78578, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2025-11-12 Classification: ISOMERASE/DNA/ISOMERASE INHIBITOR Ligands: A1L5V |
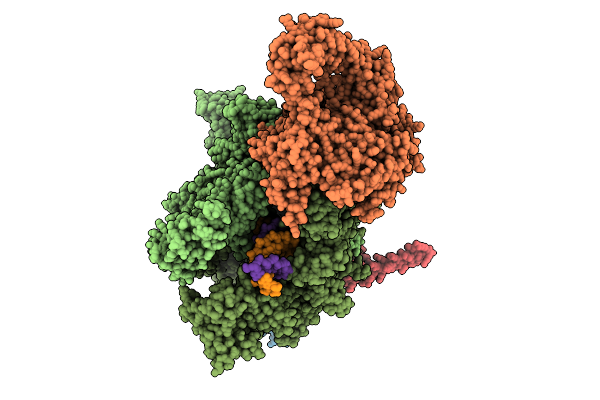 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia phage t4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: ISOMERASE Ligands: MG |
 |
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: MG, JHN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-10-08 Classification: DNA Ligands: A1L8R |
 |
Organism: Escherichia phage t4, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ISOMERASE/DNA |
 |
Human Topoisomerase 2 Alpha Atpase Domain Bound To Non-Hydrolyzable Atp Analog Amppnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-06-25 Classification: ISOMERASE Ligands: ANP, MG |

