Search Count: 13
 |
Minimal Puta Proline Dehydrogenase Domain (Design #1) Complexed With S-(-)-Tetrahydro-2-Furoic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-14 Classification: OXIDOREDUCTASE Ligands: FAD, TFB |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With S-(-)-Tetrahydro-2-Furoic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2022-12-14 Classification: OXIDOREDUCTASE Ligands: FAD, TFB |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: TFB, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: NDP, TFB, PEG |
 |
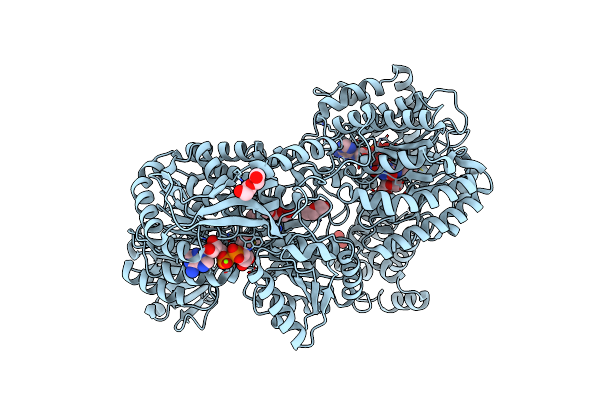
Structure Of Proline Utilization A From Sinorhizobium Meliloti Complexed With L-Tetrahydrofuroic Acid And Nad+ In Space Group P21
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-10-05 Classification: OXIDOREDUCTASE Ligands: FAD, NAD, TFB, MG, PGE, PG4 |
 |
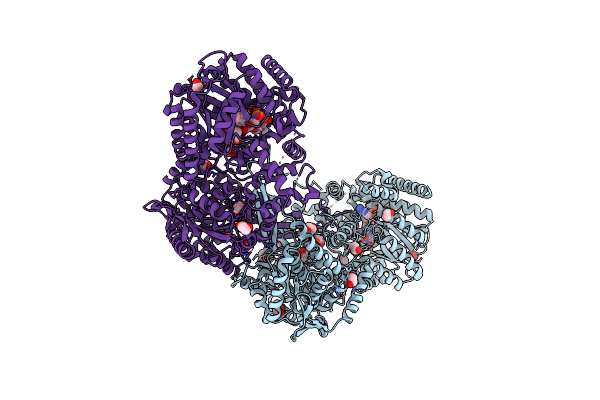
Structure Of Proline Utilization A From Sinorhizobium Meliloti Complexed With L-Tetrahydrofuroic Acid And Nad+ In Space Group P3121
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-10-05 Classification: OXIDOREDUCTASE Ligands: FAD, NAD, TFB, PGE, PG4, 1PE, SO4, MG |
 |
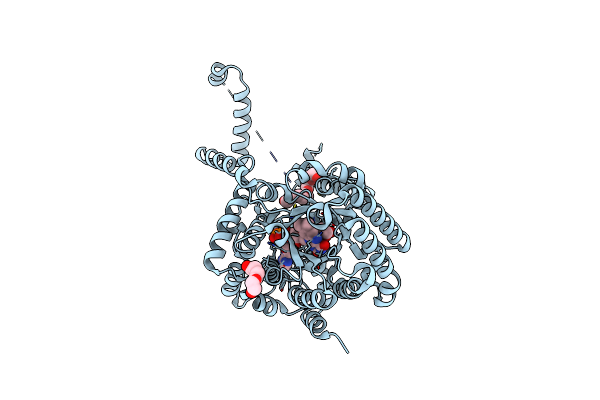
Crystal Structure Of Proline Utilization A (Puta) From Geobacter Sulfurreducens Pca In Complex With L-Tetrahydro-2-Furoic Acid
Organism: Geobacter sulfurreducens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-02-19 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, EDO |
 |
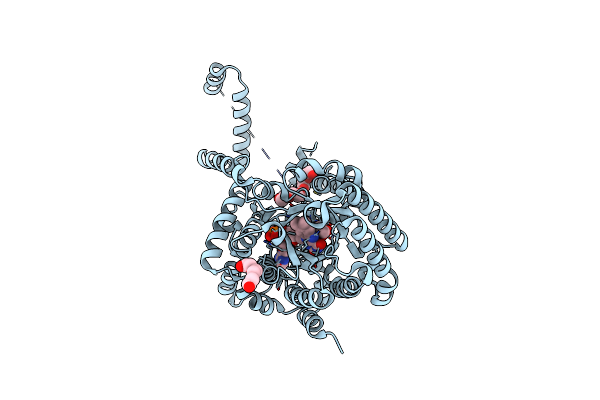
Crystal Structure Of Puta86-630 Mutant D370A Complexed With L-Tetrahydro-2-Furoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, 1PE |
 |
Crystal Structure Of Puta86-630 Mutant D370N Complexed With L-Tetrahydro-2-Furoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, 1PE |
 |
Structure Of Oxidized Deinococcus Radiodurans Proline Dehydrogenase Complexed With L-Tetrahydrofuroic Acid
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2012-11-28 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, GOL |
 |
Crystal Structure Puta86-630 Mutant Y540S Complexed With L-Tetrahydro-2-Furoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, CIT, 1PE |
 |
Crystal Structure Of E. Coli Puta Proline Dehydrogenase Domain (Residues 86-669) Complexed With L-Tetrahydro-2-Furoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-10-26 Classification: OXIDOREDUCTASE Ligands: FAD, TFB |
 |
Crystal Structure Of Quinohemoprotein Alcohol Dehydrogenase From Comamonas Testosteroni
Organism: Comamonas testosteroni
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2001-12-28 Classification: OXIDOREDUCTASE Ligands: CA, HEC, TFB, PQQ, GOL |

