Search Count: 25
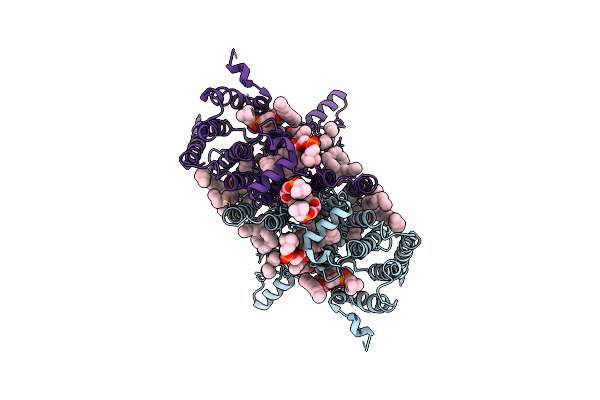 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.25 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: PSF, LBN, D12, C14, T7X |
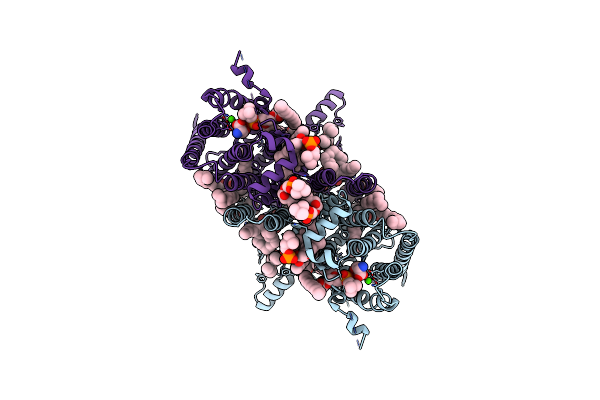 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.02 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: PSF, T7X, LBN, D12, C14, CA, SER |
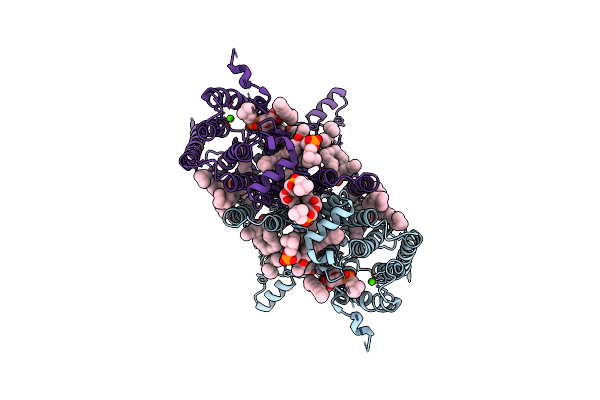 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.95 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: PSF, LBN, D12, C14, CA, T7X |
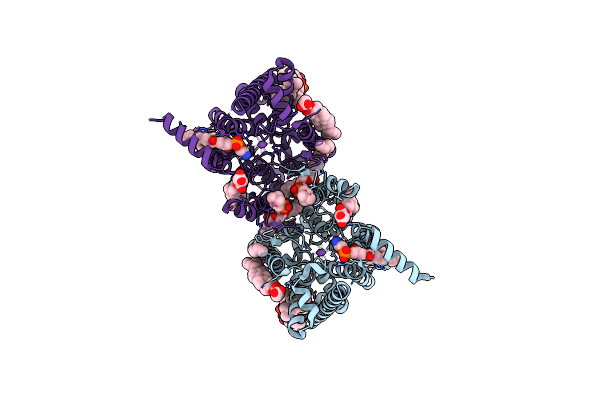 |
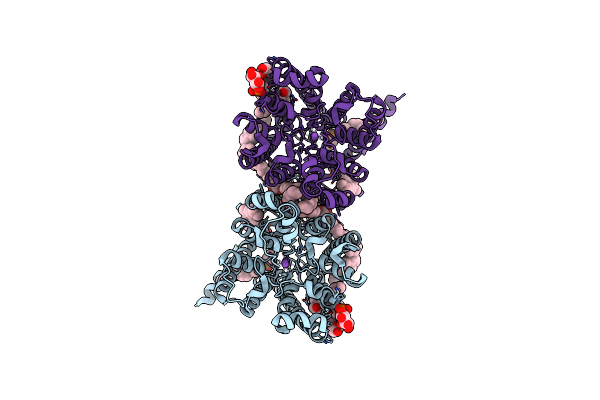
Organism: Oryza sativa subsp. japonica
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: Y01, T7X, PTY, CLR, PC1, NA |
 |
Organism: Oryza sativa subsp. indica
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: Y01, T7X, PTY, CLR, PC1, NA |
 |
Cryo-Em Structure Of The Photosystem I - Lhci Supercomplex From Coelastrella Sp.
|
 |
Cryo-Em Structure Of Tetrahymena Thermophila Respiratory Mega-Complex Mc Iv2+(I+Iii2+Ii)2
|
 |
Late Assembly Intermediate Of The Proximal Proton Pumping Module Of Complex I With Assembly Factors Ndufaf1 And Cia84
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: ELECTRON TRANSPORT Ligands: CDL, PLC, CPL, 3PE, T7X |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FMN, UQ9, PTY, PC7, LMN, NDP, ZN, 8Q1, PGT, PSF, T7X |
 |
 |
Organism: Bison bison
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: TRANSPORT PROTEIN Ligands: Y01, T7X |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2021-12-15 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FMN, UQ9, PTY, PC7, PGT, FE, NDP, ZN, 8Q1, LMN, PSF, T7X |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2021-12-15 Classification: ELECTRON TRANSPORT Ligands: SF4, PTY, FES, FMN, UQ9, LMN, FE, NDP, ZN, 8Q1, PC7, PGT, T7X, PSF |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: ELECTRON TRANSPORT Ligands: UQ9, PTY, LMN, FE, PC7, ZN, PGT, PSF, T7X |
 |
 |
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, NAI, UQ9, 3PE, PLC, CPL, T7X, CDL, NDP, LMN, ZN, ZMP |
 |
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, PLC, 3PE, LMN, T7X, CPL, CDL, NDP, ZN, ZMP |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:2.63 Å Release Date: 2021-07-28 Classification: MEMBRANE PROTEIN Ligands: T7X, LBN, 6OU, NA, YFP |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:3.06 Å Release Date: 2021-07-28 Classification: MEMBRANE PROTEIN Ligands: 6OU, LBN, YFP, T7X |
 |
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Resolution:2.96 Å Release Date: 2020-10-28 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, NDP, 3PE, LMN, ZN, ZMP, PLC, CDL, UQ9, T7X, PSC |

