Search Count: 55
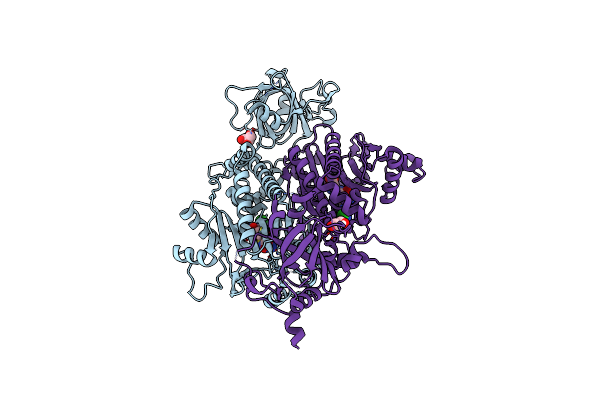 |
Structural Studies Of Reaction Hijacking Inhibition Of A Malaria Parasite Aspartyl-Trna Synthetase.
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: LIGASE Ligands: A1B0L, GOL |
 |
Organism: Yasminevirus sp. gu-2018
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-10-02 Classification: LIGASE Ligands: AMP, NA, MG |
 |
Organism: Yasminevirus sp. gu-2018
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-10-02 Classification: LIGASE Ligands: ATP |
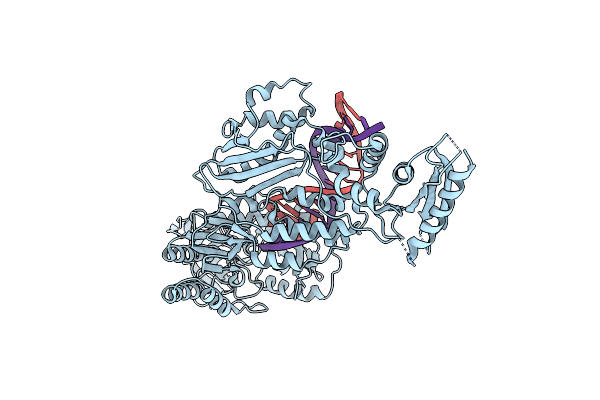 |
Structure Of The Foamy Viral Protease-Reverse Transcriptase In Complex With Rna/Dna Hybrid.
Organism: White-tufted-ear marmoset simian foamy virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-06-30 Classification: VIRAL PROTEIN Ligands: HOH |
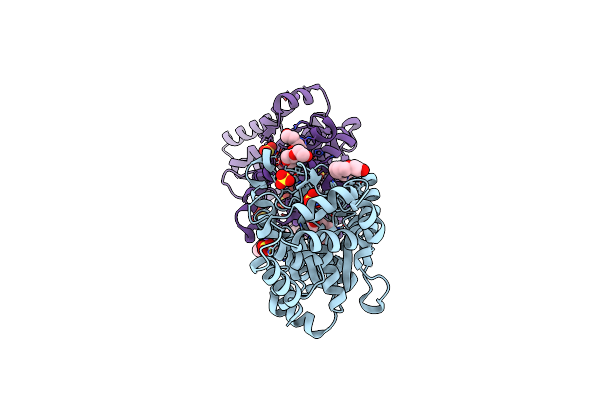 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-09-02 Classification: LIGASE Ligands: TYS, PG4, NA, SO4 |
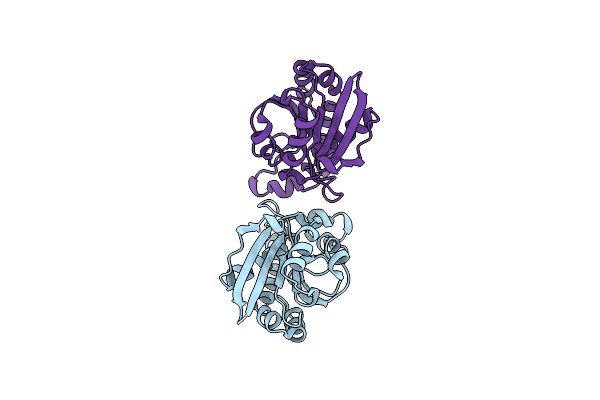 |
Crystal Structure Of A Dimeric Piptidyl T-Rna Hydrolase From Acinetobacter Baumannii At 1.50 A Resolution Reveals An Inhibited Form.
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-02-05 Classification: HYDROLASE |
 |
Crystal Structure Of Piptidyl T-Rna Hydrolase From Acinetobacter Baumannii With Bound Nacl At The Substrate Binding Site
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-08-07 Classification: HYDROLASE Ligands: CL, NA, EDO |
 |
Crystal Structure Of Ordered Asn70 And Asn116 In Native Peptidyl T-Rna Hydrolase From Acinetobacter Baumannii At 1.80 Angstrom Resolution.
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.05 Å Release Date: 2018-05-16 Classification: PROTEIN TRANSPORT/RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2018-05-16 Classification: PROTEIN TRANSPORT/RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-05-16 Classification: PROTEIN TRANSPORT/RNA BINDING PROTEIN |
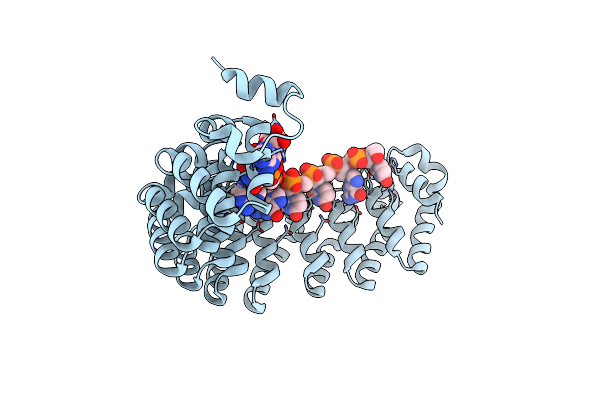 |
Crystal Structure Of The Engineered Nine-Repeat Puf Domain In Complex With Cognate 9Nt-Rna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-03-14 Classification: RNA BINDING PROTEIN/RNA |
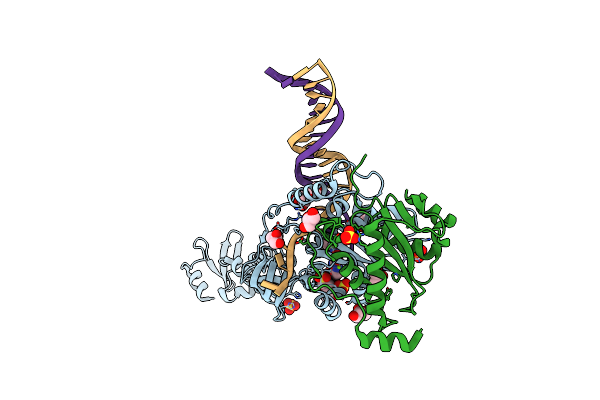 |
Crystal Structure Of The Prototype Foamy Virus (Pfv) Intasome In Complex With Magnesium And The Insti Xz384 (Compound 4A)
Organism: Human spumaretrovirus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2016-02-17 Classification: RECOMBINATION Ligands: ZN, MG, SO4, MES, GOL, WA5 |
 |
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-08-19 Classification: TRANSFERASE Ligands: LYN, TRS |
 |
Mutant K20E Of Rna Dependent Rna Polymerase 3D From Foot-And-Mouth Disease Virus Complexed With Rna
Organism: Foot-and-mouth disease virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-05-06 Classification: TRANSFERASE Ligands: MN |
 |
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-11-05 Classification: VIRAL PROTEIN/REPLICATION INHIBITOR Ligands: 20V, SO4 |
 |
Murine Norovirus Rna-Dependent-Rna-Polymerase In Complex With Compound 6, A Suramin Derivative
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-10-15 Classification: VIRAL PROTEIN/TRANSCRIPTION INHIBITOR Ligands: 2NG, MG |
 |
Crystal Structure Of Hcv Polymerase Ns5B Genotype 2A Jfh-1 Isolate With The S15G, C223H, V321I Resistance Mutations Against The Guanosine Analog Gs-0938 (Psi-3529238)
Organism: Hepatitis c virus jfh-1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-09-10 Classification: Viral Protein, transferase Ligands: MES, GOL, PG6, CL |
 |
Crystal Structure Of Human Norovirus Rna-Dependent Rna-Polymerase Bound To The Inhibitor Ppnds
Organism: Norovirus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-02-12 Classification: VIRAL PROTEIN/REPLICATION INHIBITOR/RNA Ligands: 20V, MG |
 |
Crystal Structure Of Human Norovirus Rna-Dependent Rna-Polymerase In Complex With Naf2
Organism: Norovirus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2014-02-12 Classification: VIRAL PROTEIN/REPLICATION INHIBITOR Ligands: 21D, MG |

