Search Count: 7
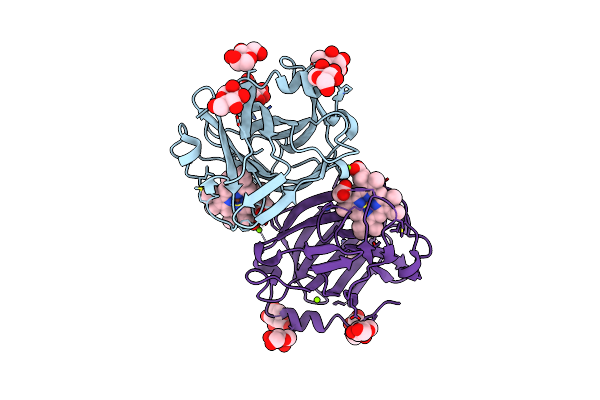 |
Cytochrome Domain Of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt
Organism: Myriococcum thermophilum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: HEM, NAG, MAN, MG |
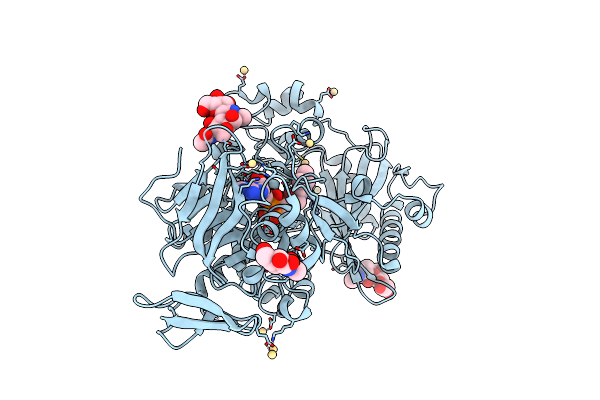 |
Dehydrogenase Domain Of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtdh
Organism: Myriococcum thermophilum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, CD |
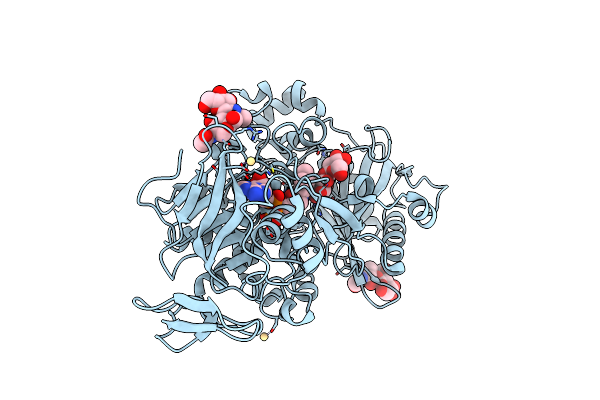 |
Dehydrogenase Domain Of Myriococcum Thermophilum Cellobiose Dehydrogenase With Bound Cellobionolactam, Mtdh
Organism: Myriococcum thermophilum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: FAD, ABL, CD |
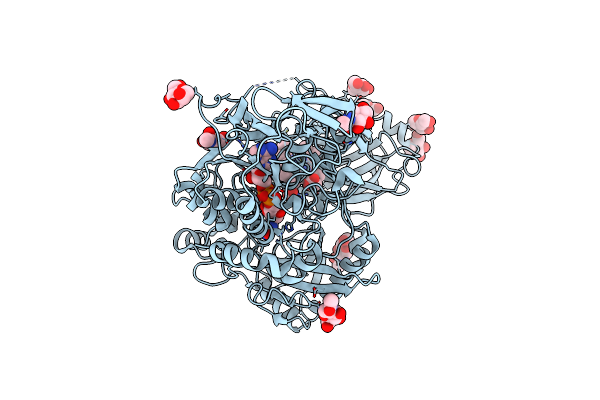 |
Organism: Myriococcum thermophilum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: HEM, FAD, NAG, MAN |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: HEM, FAD, NAG, MAN, PT, MG |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: CU, NO3 |
 |
Crystal Structure Of Pyranose Dehydrogenase From Agaricus Meleagris, Wildtype
Organism: Leucoagaricus meleagris
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: FED, NAG, PO4 |

