Search Count: 5
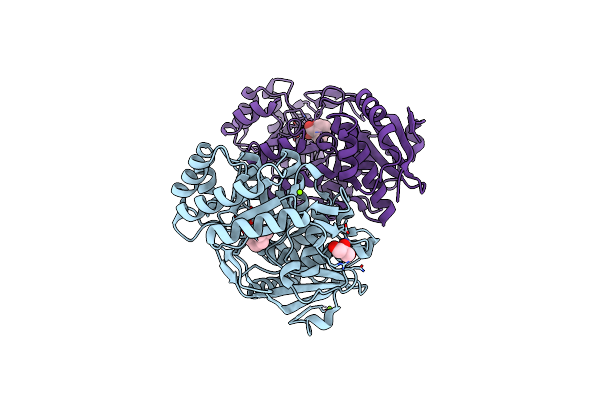 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans Pd1222 (Target Nysgrc-012907) With Bound L-Proline Betaine (Substrate)
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-03-06 Classification: ISOMERASE Ligands: MG, IOD, GOL, PBE |
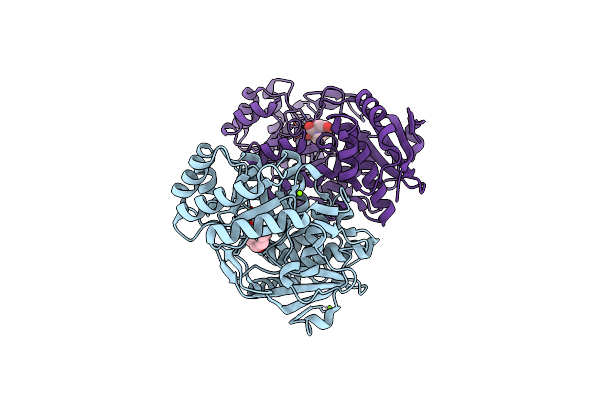 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans Pd1222 (Target Nysgrc-012907) With Bound Cis-4Oh-D-Proline Betaine (Product)
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: IOD, MG, 4OP |
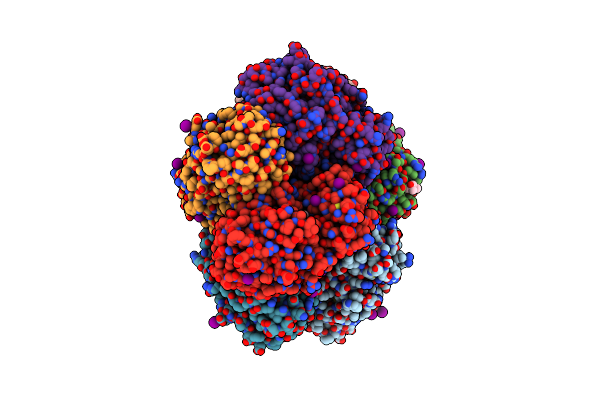 |
Crystal Structure Of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis Htcc2601, With Bound Mg And L-4-Hydroxyproline Betaine (Betonicine)
Organism: Pelagibaca bermudensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-10 Classification: ISOMERASE Ligands: MG, NI, 0XW, IOD, MPD |
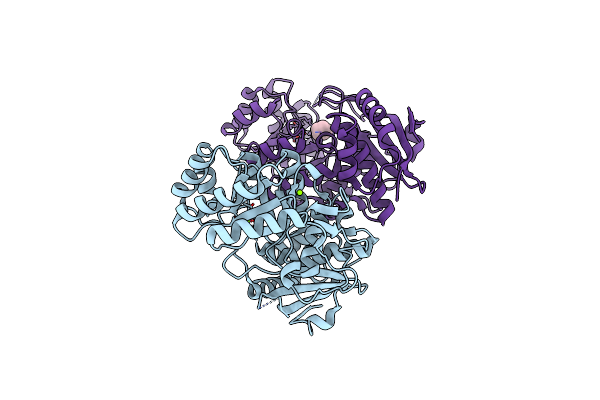 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans Pd1222 (Target Nysgrc-012907) With Bound Mg
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-05-02 Classification: ISOMERASE Ligands: MG, UNL |
 |
Crystal Structure Of A Mandelate Racemase/Muconate Lactonizing Enzyme From Roseovarius Sp. Htcc2601
Organism: Roseovarius sp.
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2007-05-08 Classification: ISOMERASE Ligands: MG |

