Search Count: 41
 |
Jacalin-Carbohydrate Interactions. Distortion Of The Ligand As A Determinant Of Affinity
Organism: Artocarpus integer
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2015-02-18 Classification: SUGAR BINDING PROTEIN Ligands: MBG, IPA, EDO |
 |
Jacalin-Carbohydrate Interactions. Distortion Of The Ligand As A Determinant Of Affinity.
Organism: Artocarpus integer
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-02-18 Classification: SUGAR BINDING PROTEIN Ligands: ZZ1, GLA, IPA, EDO |
 |
Jacalin-Carbohydrate Interactions. Distortion Of The Ligand As A Determinant Of Affinity.
Organism: Artocarpus integer
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-02-18 Classification: SUGAR BINDING PROTEIN Ligands: IPA, EDO, ZZ1, GAL |
 |
Jacalin-Carbohydrate Interactions. Distortion Of The Ligand As A Determinant Of Affinity.
Organism: Artocarpus integer
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-02-18 Classification: SUGAR BINDING PROTEIN Ligands: NBZ, GLA, EDO, IPA |
 |
Jacalin-Carbohydrate Interactions. Distortion Of The Ligand As A Determinant Of Affinity.
Organism: Artocarpus integer
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2015-02-18 Classification: SUGAR BINDING PROTEIN Ligands: 147, EDO, IPA |
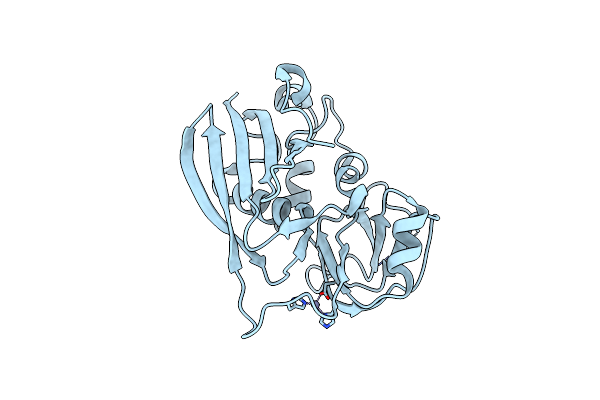 |
Manipulating The Coupled Folding And Binding Process Drives Affinity Maturation In A Protein-Protein Complex
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-05-12 Classification: TOXIN Ligands: ZN |
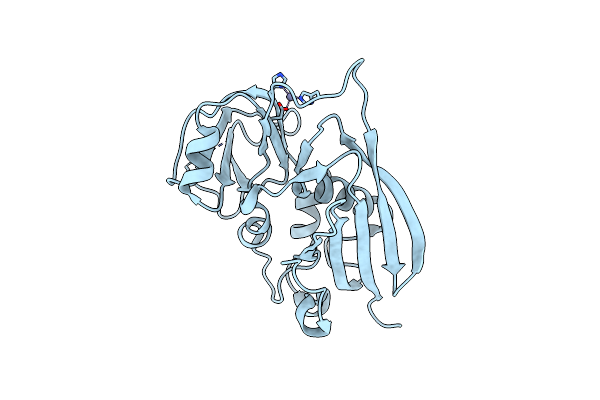 |
Manipulating The Coupled Folding And Binding Process Drives Affinity Maturation In A Protein-Protein Complex
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-05-12 Classification: TOXIN Ligands: ZN |
 |
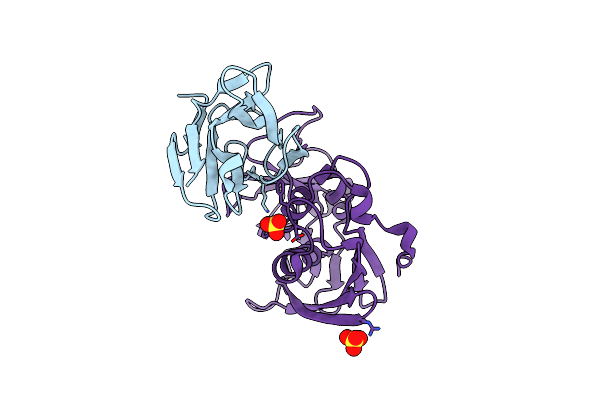
A Complex Between A Variant Of Staphylococcal Enterotoxin C3 And The Variable Domain Of The Murine T Cell Receptor Beta Chain 8.2
Organism: Mus musculus, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-05-12 Classification: TOXIN |
 |
Manipulating The Coupled Folding And Binding Process Drives Affinity Maturation In A Protein-Protein Complex
Organism: Mus musculus, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-05-12 Classification: TOXIN Ligands: SO4 |
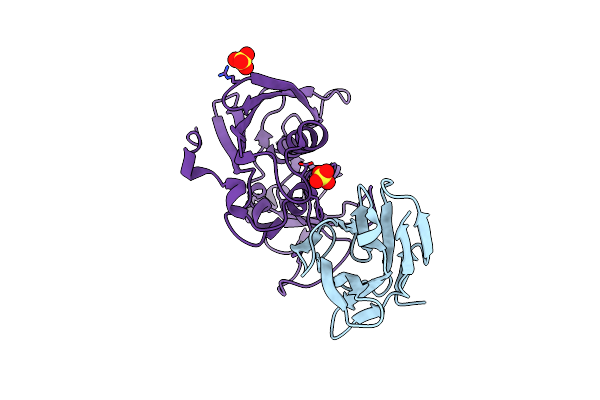 |
Manipulating The Coupled Folding And Binding Process Drives Affinity Maturation In A Protein-Protein Complex
Organism: Mus musculus, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-05-12 Classification: TOXIN Ligands: SO4 |
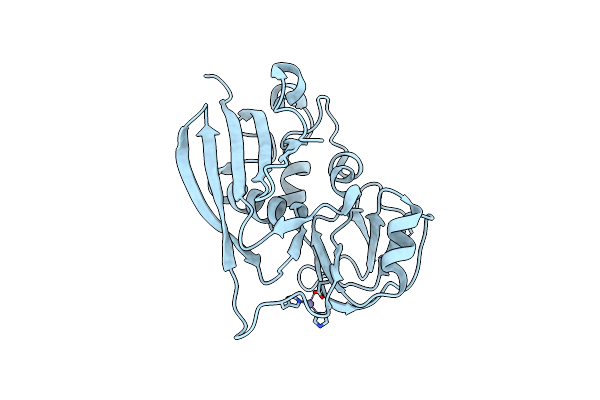 |
Manipulating The Coupled Folding And Binding Process Drives Affinity Maturation In A Protein-Protein Complex
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-01-27 Classification: TOXIN Ligands: ZN |
 |
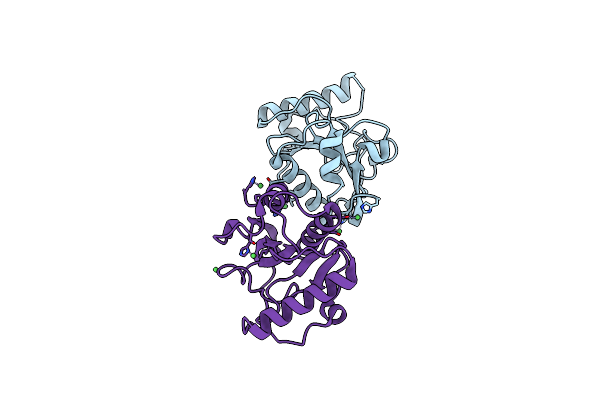
Crystal Structure Of Human Pgrp-Ibetac In Complex With Glycosamyl Muramyl Pentapeptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-10-02 Classification: PEPTIDOGLYCAN-BINDING PROTEIN |
 |
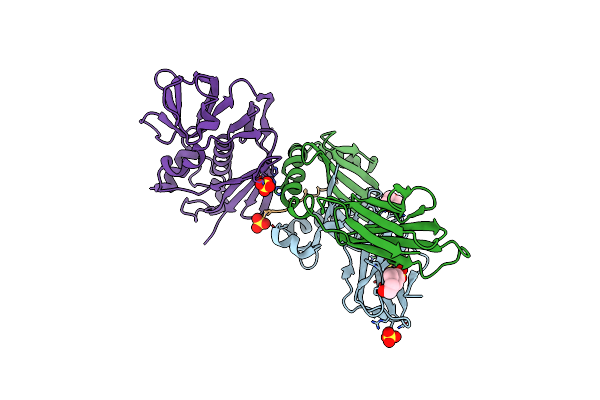
Crystal Structure Of The C-Terminal Peptidoglycan-Binding Domain Of Human Peptidoglycan Recognition Protein Ibeta
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-09-18 Classification: SUGAR BINDING PROTEIN Ligands: NI |
 |
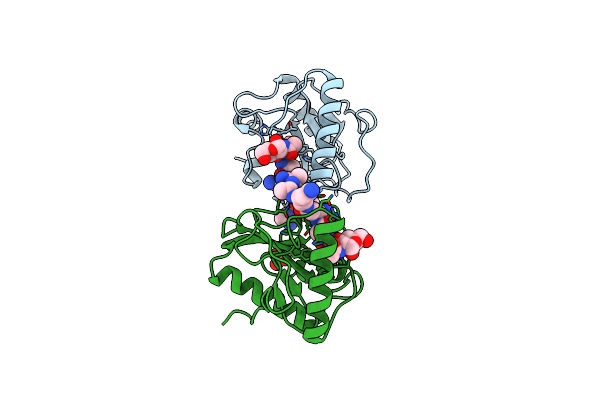
Crystal Structure Of Staphylococcal Enterotoxin I (Sei) In Complex With A Human Mhc Class Ii Molecule
Organism: Homo sapiens, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-07-11 Classification: IMMUNE SYSTEM Ligands: SO4, EPE, DIO, ZN |
 |
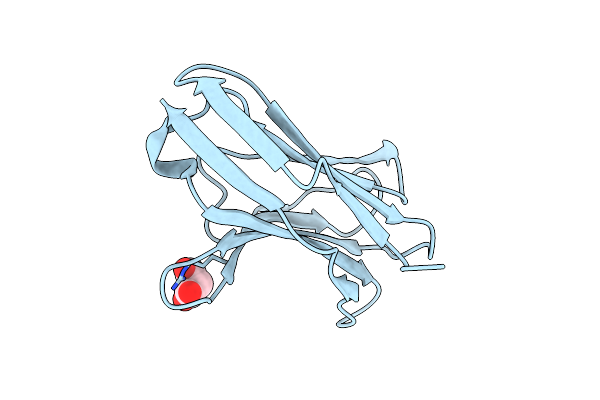
Crystal Structure Of Human Pgrp-Ialphac In Complex With Muramyl Pentapeptide
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-06-27 Classification: IMMUNE SYSTEM Ligands: SO4, AMU |
 |
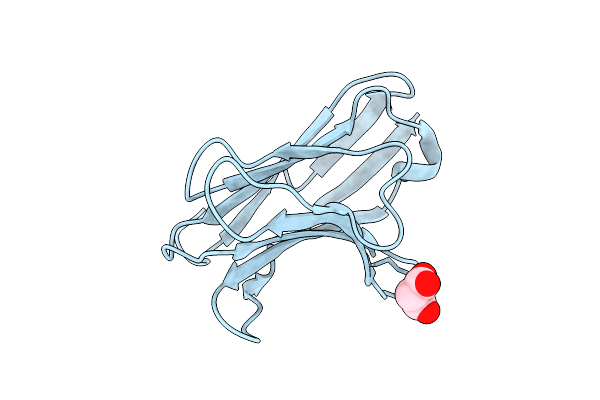
Crystal Structure Of The S54N Variant Of Murine T Cell Receptor Vbeta 8.2 Domain
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-03-21 Classification: IMMUNE SYSTEM Ligands: MLA |
 |
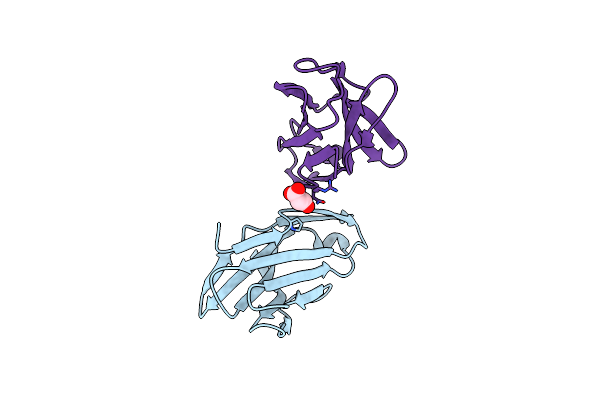
Crystal Structure Of The A52V/S54N/K66E Variant Of The Murine T Cell Receptor V Beta 8.2 Domain
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-03-21 Classification: IMMUNE SYSTEM Ligands: MLA |
 |
Crystal Structure Of The G17E/S54N/K66E/Q72H/E80V/L81S/T87S/G96V Variant Of The Murine T Cell Receptor V Beta 8.2 Domain
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-03-21 Classification: IMMUNE SYSTEM Ligands: MLA |
 |
Crystal Structure Of The G17E/A52V/S54N/Q72H/E80V/L81S/T87S/G96V Variant Of The Murine T Cell Receptor V Beta 8.2 Domain
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-03-21 Classification: IMMUNE SYSTEM Ligands: MLA |
 |
Crystal Structure Of The G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V Variant Of The Murine T Cell Receptor V Beta 8.2 Domain
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-03-21 Classification: IMMUNE SYSTEM Ligands: MLA |

