Search Count: 18
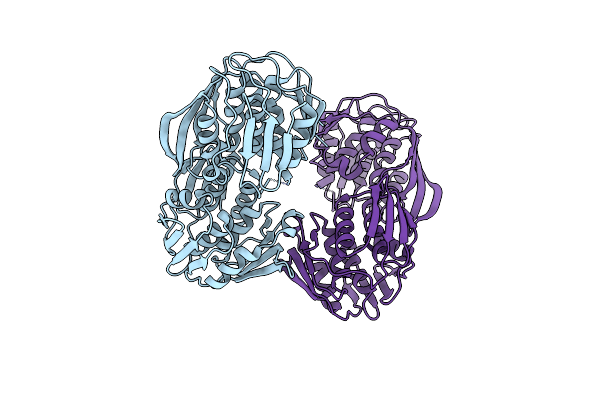 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2016-02-24 Classification: TRANSFERASE |
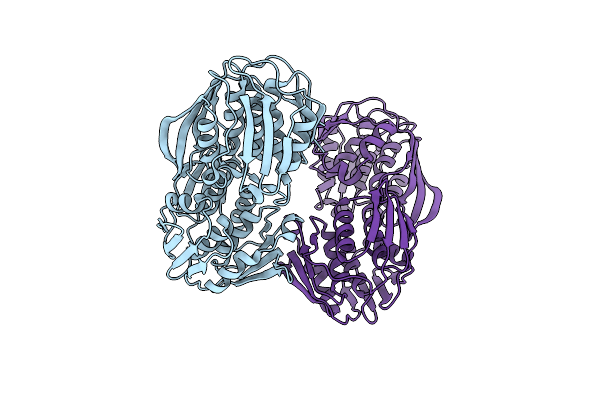 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: CL |
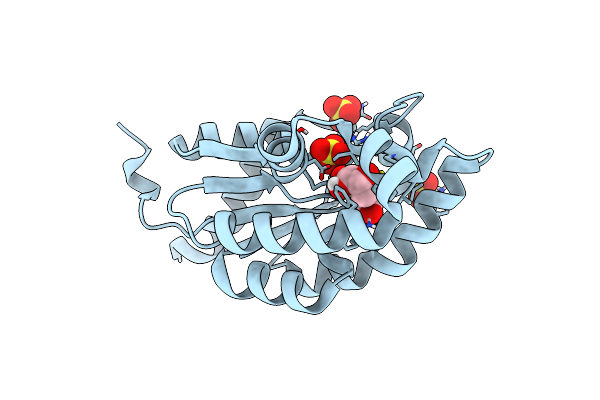 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2015-08-12 Classification: transferase/transferase inhibitor Ligands: SKM, SO4 |
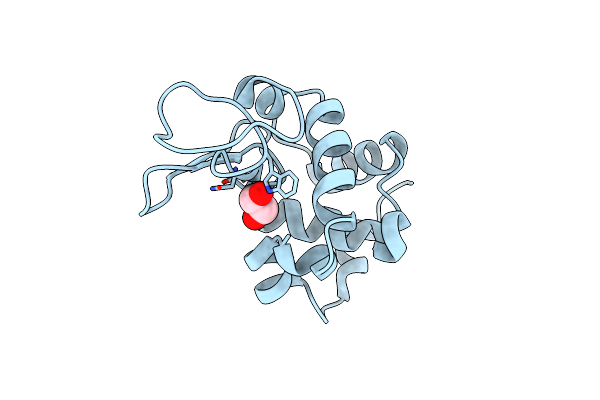 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, EDO |

