Search Count: 30
 |
Organism: Hepatitis b virus genotype d subtype adw (isolate united kingdom/adyw/1979)
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: A1BVU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2Q |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2T |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2Q |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2R, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: TRANSFERASE Ligands: A1AF6 |
 |
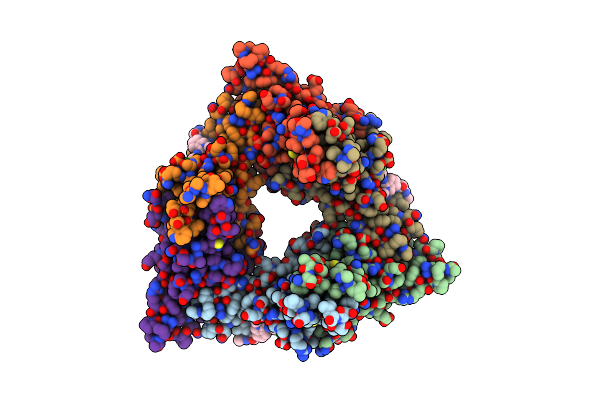
Organism: Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-09-25 Classification: VIRAL PROTEIN Ligands: A1AVK |
 |
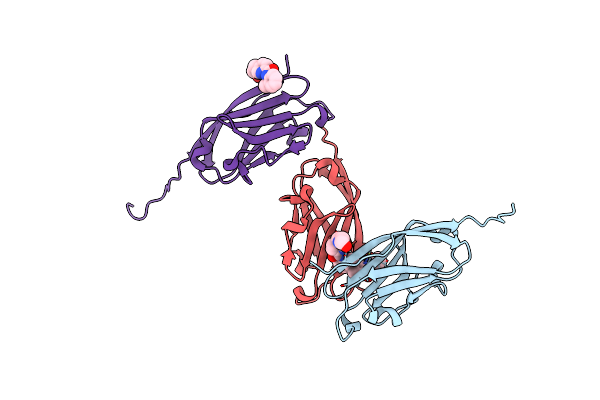
Organism: Hepatitis b virus genotype a
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-05-04 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 8EI |
 |
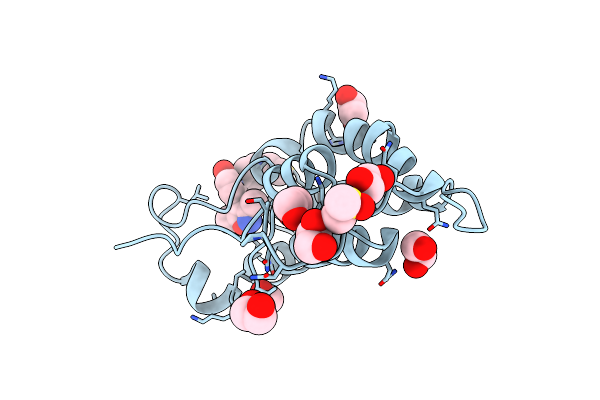
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-01-20 Classification: CELL CYCLE Ligands: R81 |
 |
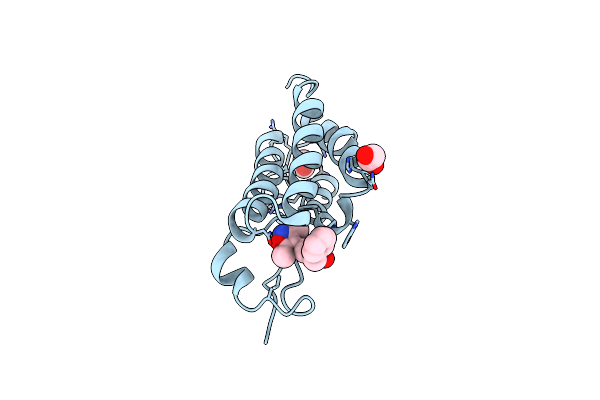
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2018-09-26 Classification: TRANSCRIPTION/INHIBITOR Ligands: FP7, EDO, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-09-26 Classification: TRANSCRIPTION/INHIBITOR Ligands: FOY, EDO |
 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-09-05 Classification: RNA Ligands: MG, GZ7, K |
 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2018-09-05 Classification: RNA Ligands: MG, GZG, K |
 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-05 Classification: RNA Ligands: CL, MG, GZ4, K |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Follow On Fragment Ebsi-4723 4-(5-Chlorothiophen-2-Yl)-1H-Pyrazole
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-03-08 Classification: TRANSCRIPTION Ligands: 8QG |
 |
X-Ray Crystal Structure Of Human Brd2(Bd2) In Complex With Rvx297 To 1.55 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2016-06-22 Classification: PROTEIN BINDING/INHIBITOR Ligands: 5GD, NA |

