Search Count: 23
 |
Organism: Methanopyrus kandleri av19
Method: SOLUTION NMR Release Date: 2019-02-13 Classification: UNKNOWN FUNCTION |
 |
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-11-28 Classification: PLANT PROTEIN |
 |
Domain Swapped Dimer Of Engineered Hairpin Loop1 Mutant In Single-Chain Monellin
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2018-11-28 Classification: PLANT PROTEIN |
 |
Double Domain Swapped Dimer Of Engineered Hairpin Loop1 And Loop3 Mutant In Single-Chain Monellin
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2018-11-28 Classification: PLANT PROTEIN |
 |
Organism: Caenorhabditis elegans
Method: SOLUTION NMR Release Date: 2017-11-08 Classification: SIGNALING PROTEIN |
 |
Domain Swapped Dimer Crystal Structure Of Loop1 Deletion Mutant In Single-Chain Monellin
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2017-07-26 Classification: PLANT PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-05-18 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-05-18 Classification: PROTEIN BINDING |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-01-14 Classification: TRANSFERASE/DNA Ligands: MG, 1FZ |
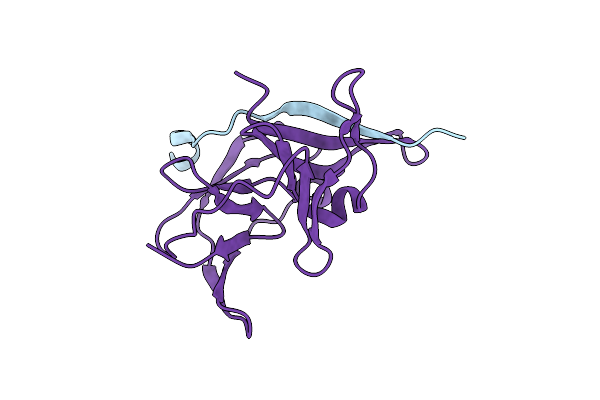 |
Organism: Japanese encephalitis virus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: CL |
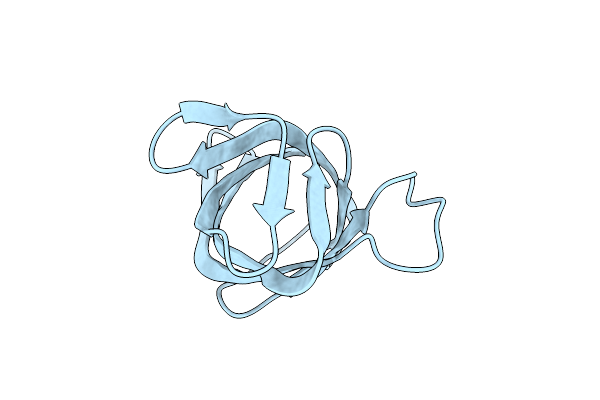 |
Crystal Structure Of Hypothetical Protein Pf0907 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: UNKNOWN FUNCTION Ligands: CL |
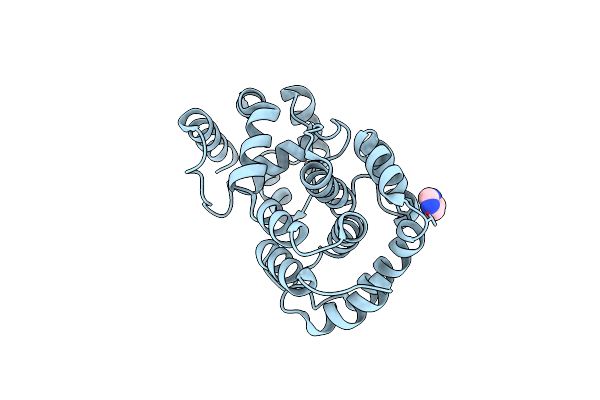 |
Crystal Structure Of Hypothetical Protein Pf0907 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-12-10 Classification: UNKNOWN FUNCTION Ligands: CL, IMD |
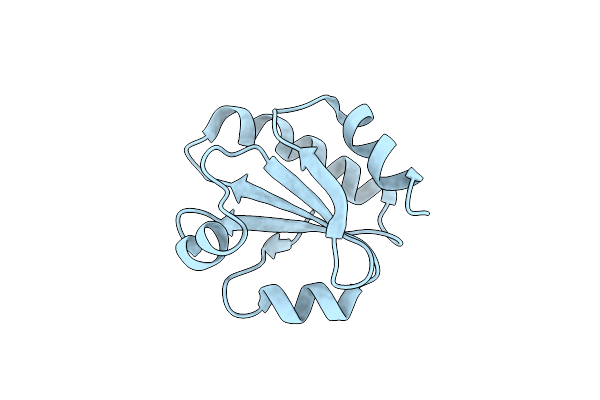 |
Crystal Structure Of Thermus Thermophilus Thioredoxin Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-12-10 Classification: ELECTRON TRANSPORT Ligands: CL |
 |
Hypothetical Protein Pf1117 From Pyrococcus Furiosus: Structure Solved By Sulfur-Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-12-10 Classification: ISOMERASE Ligands: GSH, LVJ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-12-10 Classification: CELL CYCLE |
 |
Crystal Structure Of The Exonuclease Domain Of Qip (Qde-2 Interacting Protein) Solved By Native-Sad Phasing.
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-12-10 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Conserved Hypothetical Protein Pf1771 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Japanese encephalitis virus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: GTP, ZN |
 |
Organism: Japanese encephalitis virus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: ATP, ZN |

