Search Count: 24
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-04-19 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-19 Classification: CHAPERONE, HYDROLASE Ligands: FMT |
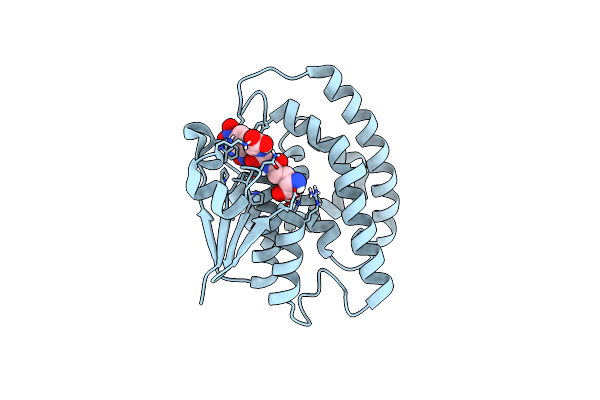 |
Organism: Alopecurus myosuroides
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: V7K |
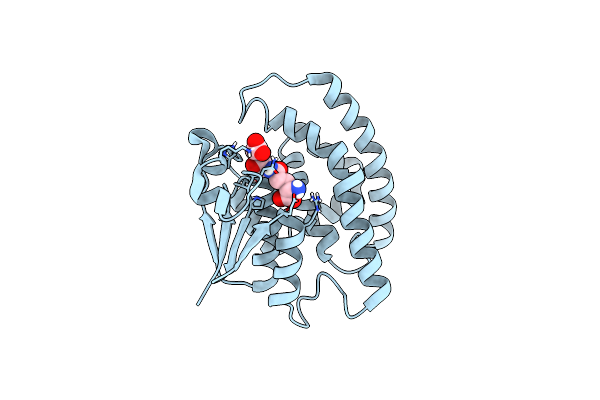 |
Organism: Alopecurus myosuroides
Method: X-RAY DIFFRACTION Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: GSH |
 |
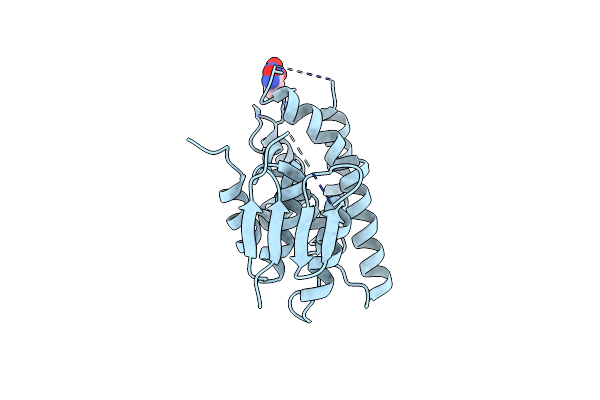
Organism: Leishmania mexicana mhom/gt/2001/u1103
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2022-02-16 Classification: CHAPERONE Ligands: GOL |
 |
Organism: Alopecurus myosuroides
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-08-18 Classification: TRANSFERASE Ligands: 5Z8 |
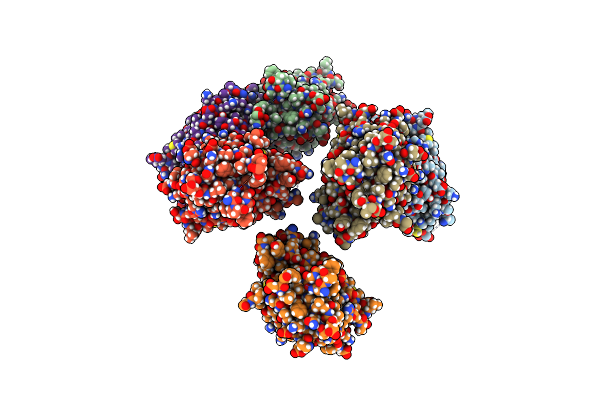 |
Organism: Alopecurus myosuroides
Method: X-RAY DIFFRACTION Release Date: 2021-06-30 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Alopecurus myosuroides
Method: X-RAY DIFFRACTION Release Date: 2021-05-26 Classification: TRANSFERASE Ligands: GSH |
 |
Organism: Alopecurus myosuroides
Method: X-RAY DIFFRACTION Release Date: 2021-05-12 Classification: TRANSFERASE |
 |
Organism: Shewanella oneidensis (strain mr-1)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2019-09-18 Classification: SIGNALING PROTEIN Ligands: GDP |
 |
Organism: Shewanella oneidensis (strain mr-1)
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2019-09-18 Classification: SIGNALING PROTEIN Ligands: GCP, GDP |
 |
Organism: Shewanella oneidensis (strain mr-1)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-09-18 Classification: SIGNALING PROTEIN Ligands: GDP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2019-01-02 Classification: CHAPERONE Ligands: ATP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:9.30 Å Release Date: 2019-01-02 Classification: CHAPERONE Ligands: ATP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2019-01-02 Classification: CHAPERONE Ligands: ATP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-01-03 Classification: CHAPERONE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2017-11-29 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2016-08-10 Classification: CHAPERONE Ligands: ANP, PO4, MG, GOL, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2016-03-02 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-03-02 Classification: CHAPERONE Ligands: ANP, MG |

