Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-03-15 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Crystal Structure Of A Novel Putative Sugar Isomerase From The Psychrophilic Bacterium Paenibacillus Sp. R4
Organism: Paenibacillus sp. fsl h7-0331
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2021-12-29 Classification: ISOMERASE Ligands: CA, ZN |
 |
Crystal Structure Of Arabinose Isomerase From Hyper Thermophilic Bacterium Thermotoga Maritima (Tmai) Triple Mutant (K264A, E265A, K266A)
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2021-10-20 Classification: ISOMERASE Ligands: MN |
 |
Organism: Alicyclobacillus sp. tp-7, Geobacillus kaustophilus (strain hta426), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2021-10-20 Classification: ISOMERASE Ligands: MN, NA |
 |
Organism: Alicyclobacillus sp. tp-7, Geobacillus kaustophilus (strain hta426), Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-09-08 Classification: ISOMERASE Ligands: GOL, MN |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-09-08 Classification: ISOMERASE Ligands: MPD, MN, RB0 |
 |
Crystal Structure Of Arabinose Isomerase From Hyper Thermophilic Bacterium Thermotoga Maritima (Tmai) Wt
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2021-09-01 Classification: ISOMERASE Ligands: MN, GOL |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-09-01 Classification: ISOMERASE Ligands: MPD, MN, GOL |
 |
Structural Insights Into Novel Mechanisms Of Inhibition Of The Major B-Carbonic Anhydrase Cafb From The Pathogenic Fungus Aspergillus Fumigatus (C116 Flipped Form)
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-03-31 Classification: LYASE Ligands: ACT |
 |
Structural Insights Into Novel Mechanisms Of Inhibition Of The Major B-Carbonic Anhydrase Cafb From The Pathogenic Fungus Aspergillus Fumigatus (Disulfide-Bonded Form)
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-03-31 Classification: LYASE Ligands: ACT, SO4 |
 |
Structural Insights Into Novel Mechanisms Of Inhibition Of The Major B-Carbonic Anhydrase Cafb From The Pathogenic Fungus Aspergillus Fumigatus (Zinc-Bound Form)
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-03-31 Classification: LYASE Ligands: ZN |
 |
Crystal Structure Of Pyrrolidone Carboxyl Peptidase From Thermophilic Keratin Degrading Bacterium Fervidobacterium Islandicum Aw-1 (Fipcp)
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: GOL, 1PE |
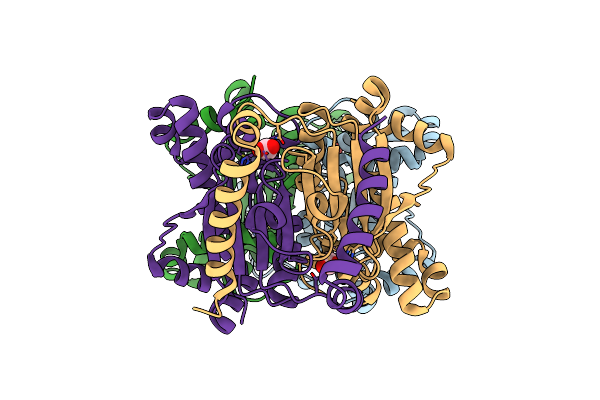 |
Crystal Structure Of The B-Carbonic Anhydrase Cafa Of The Fungal Pathogen Aspergillus Fumigatus
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-10-14 Classification: LYASE Ligands: ACT, ZN |
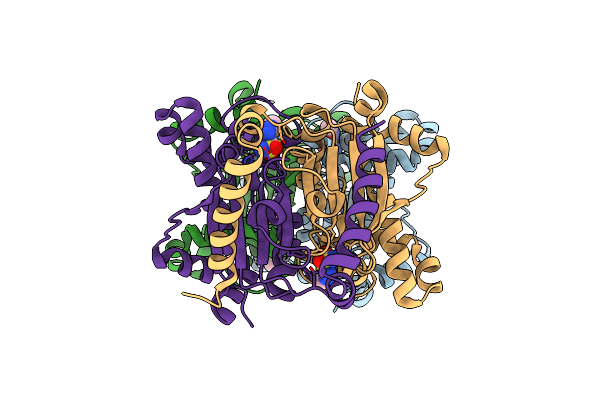 |
Crystal Structure Of The B-Carbonic Anhydrase Cafa Of The Fungal Pathogen Aspergillus Fumigatus
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: LYASE Ligands: ZN, AZM |
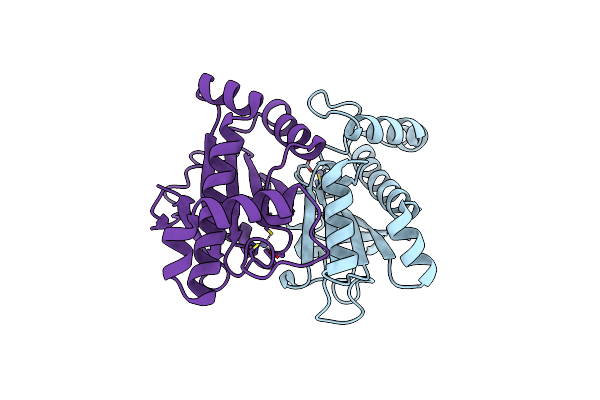 |
The Structural Basis Of The Beta-Carbonic Anhydrase Cafd (C39A Mutant) Of The Filamentous Fungus Aspergillus Fumigatus
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-11-27 Classification: LYASE Ligands: ZN |
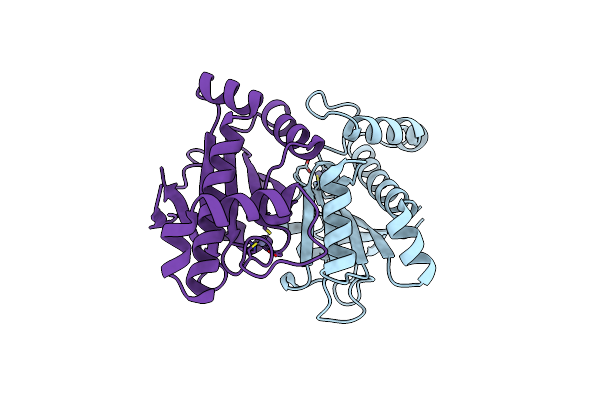 |
The Structural Basis Of The Beta-Carbonic Anhydrase Cafd (E54A Mutant) Of The Filamentous Fungus Aspergillus Fumigatus
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-27 Classification: LYASE Ligands: ZN |
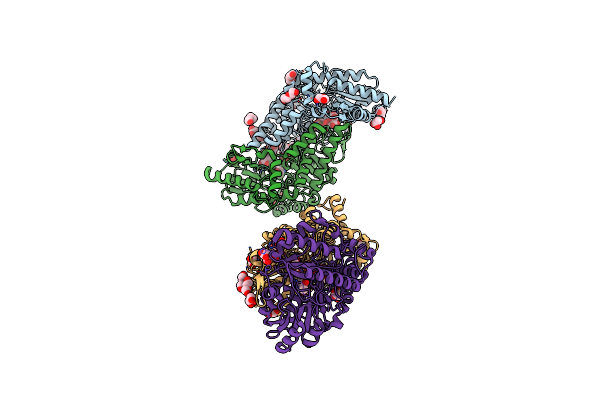 |
Crystal Structure Of Thermostable Cysteine Desulfurase (Fisufs) From Thermophilic Fervidobacterium Islandicum Aw-1
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2019-10-09 Classification: TRANSFERASE Ligands: PLP, GOL, CIT, PEG |
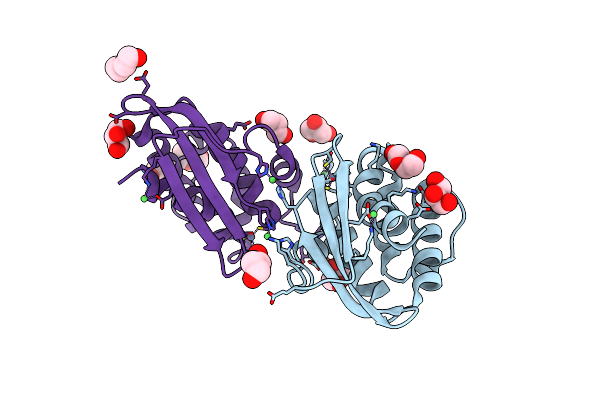 |
Crystal Structure Of Putative Iron-Sulfur Cluster Assembly Scaffold Protein For Suf System (Fisufu) From Thermophilic Fervidobacterium Islandicum Aw-1
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-09 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, PEG, ZN, NI |
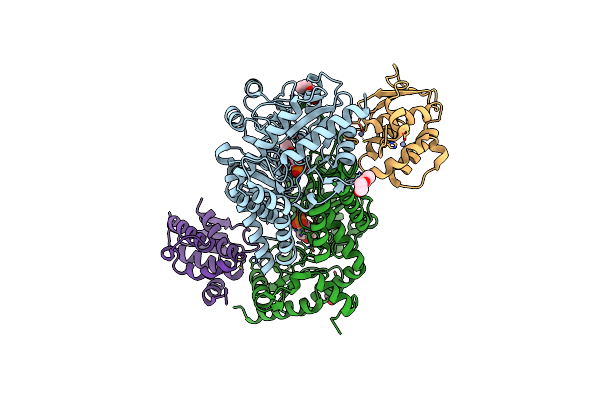 |
Crystal Structure Of Thermostable Fisufs-Sufu Complex From Thermophilic Fervidobacterium Islandicum Aw-1
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-10-09 Classification: TRANSFERASE Ligands: PLP, PEG, GOL, ZN |