Search Count: 10
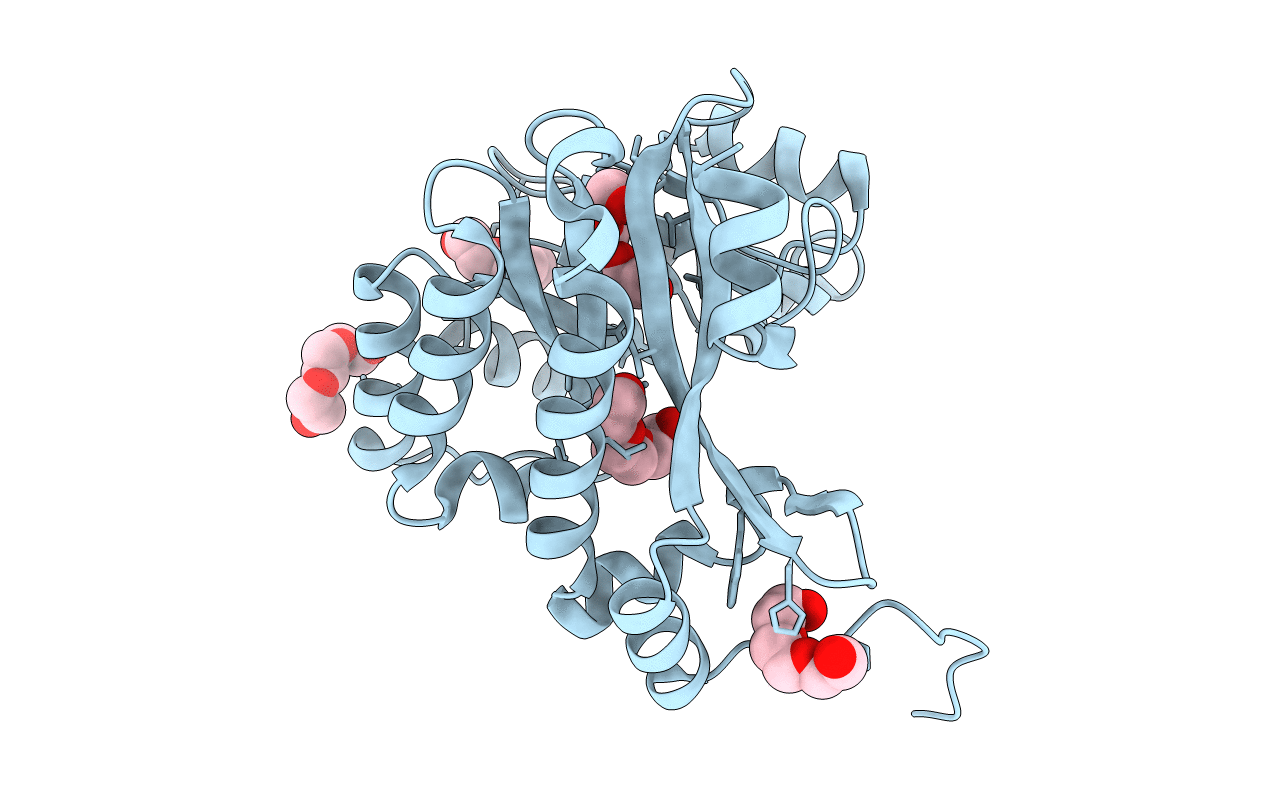 |
Crystal Structure Of The Eal Domain Of C-Di-Gmp Specific Phosphodiesterase Yaha
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-01-29 Classification: HYDROLASE Ligands: PGE |
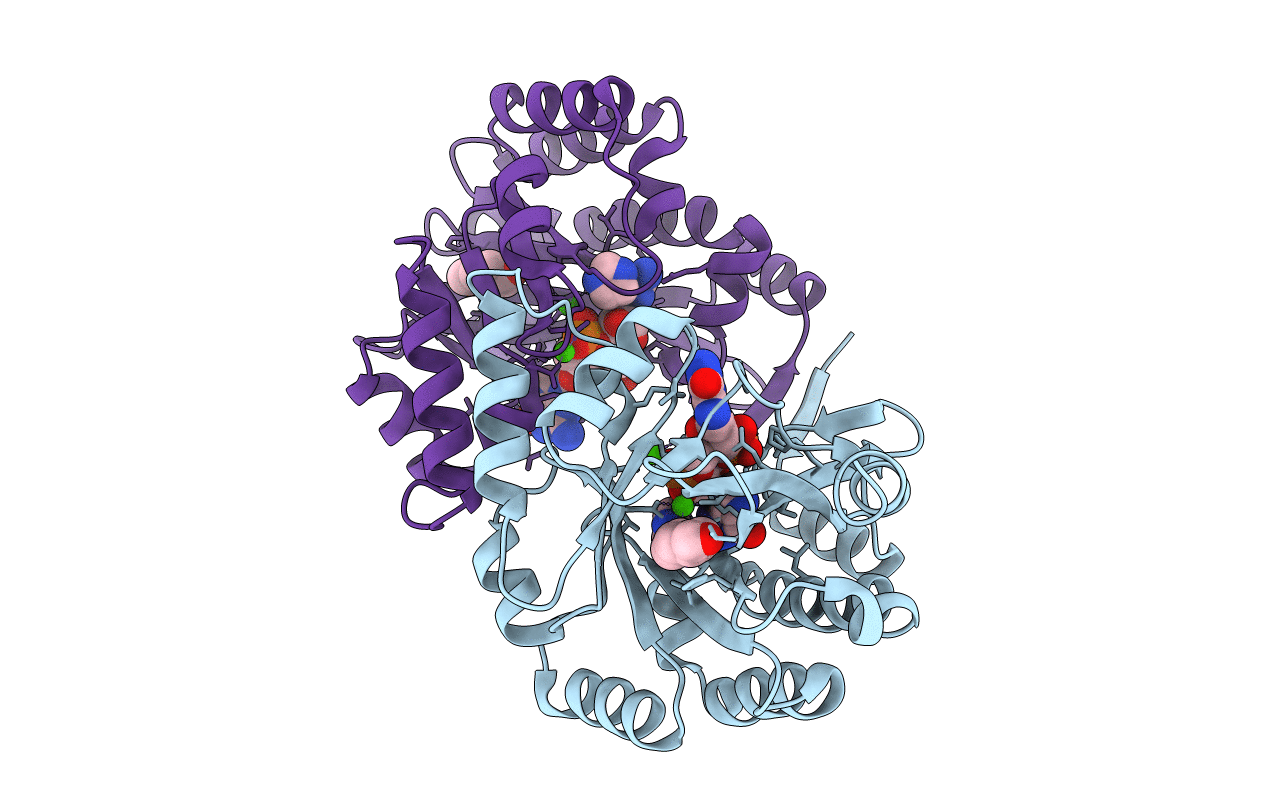 |
Crystal Structure Of The Eal Domain Of C-Di-Gmp Specific Phosphodiesterase Yaha In Complex With Substrate C-Di-Gmp And Ca++
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-01-29 Classification: HYDROLASE Ligands: C2E, CA, IMD, HB0 |
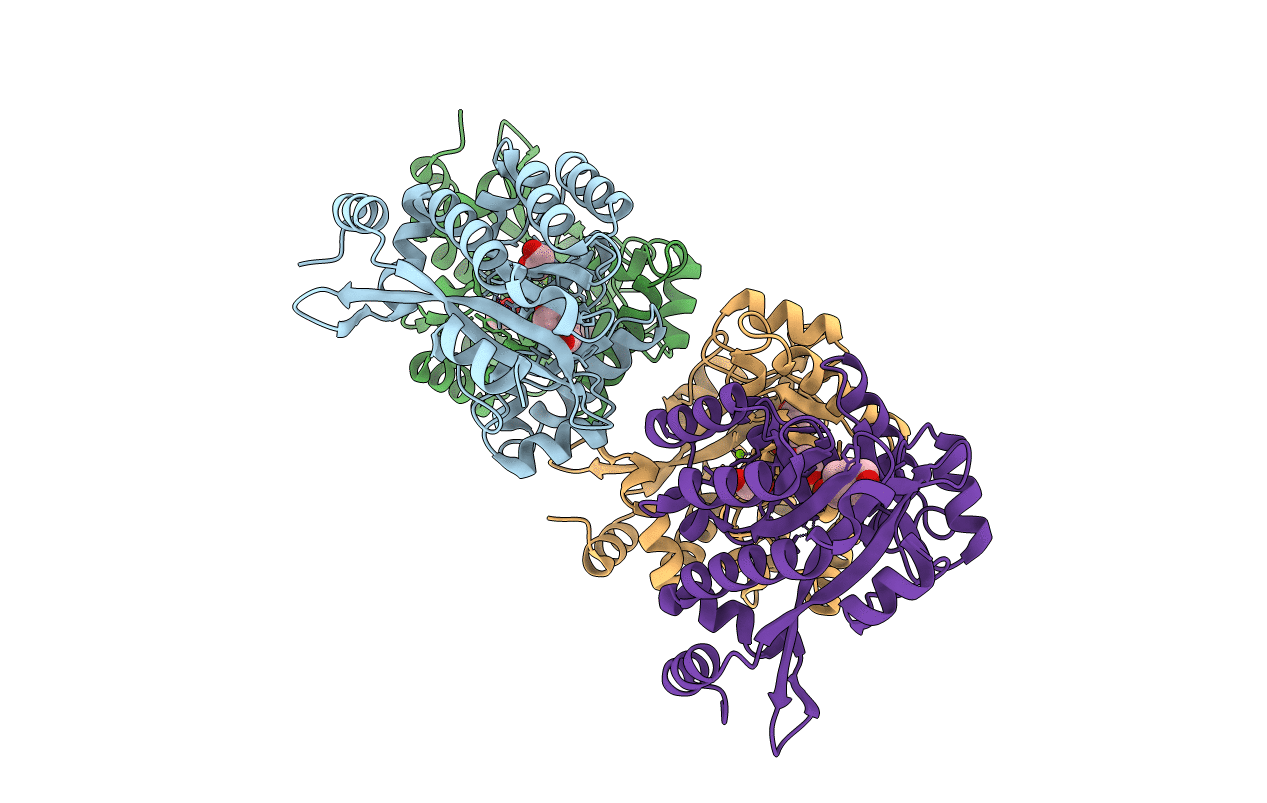 |
Crystal Structure Of The Eal Domain Of C-Di-Gmp Specific Phosphodiesterase Yaha In Complex With Activating Cofactor Mg++
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-01-29 Classification: HYDROLASE Ligands: MG, HB0, EDO |
 |
Molecular Structure Of A Metal-Independent Bacterial Glycosyltransferase That Catalyzes The Synthesis Of Histo-Blood Group A Antigen
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-12-19 Classification: TRANSFERASE |
 |
Molecular Structure Of A Metal-Independent Bacterial Glycosyltransferase That Catalyzes The Synthesis Of Histo-Blood Group A Antigen
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2012-12-19 Classification: TRANSFERASE Ligands: CA, EPE, CL |
 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-08-18 Classification: TRANSFERASE |
 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-08-18 Classification: TRANSFERASE |
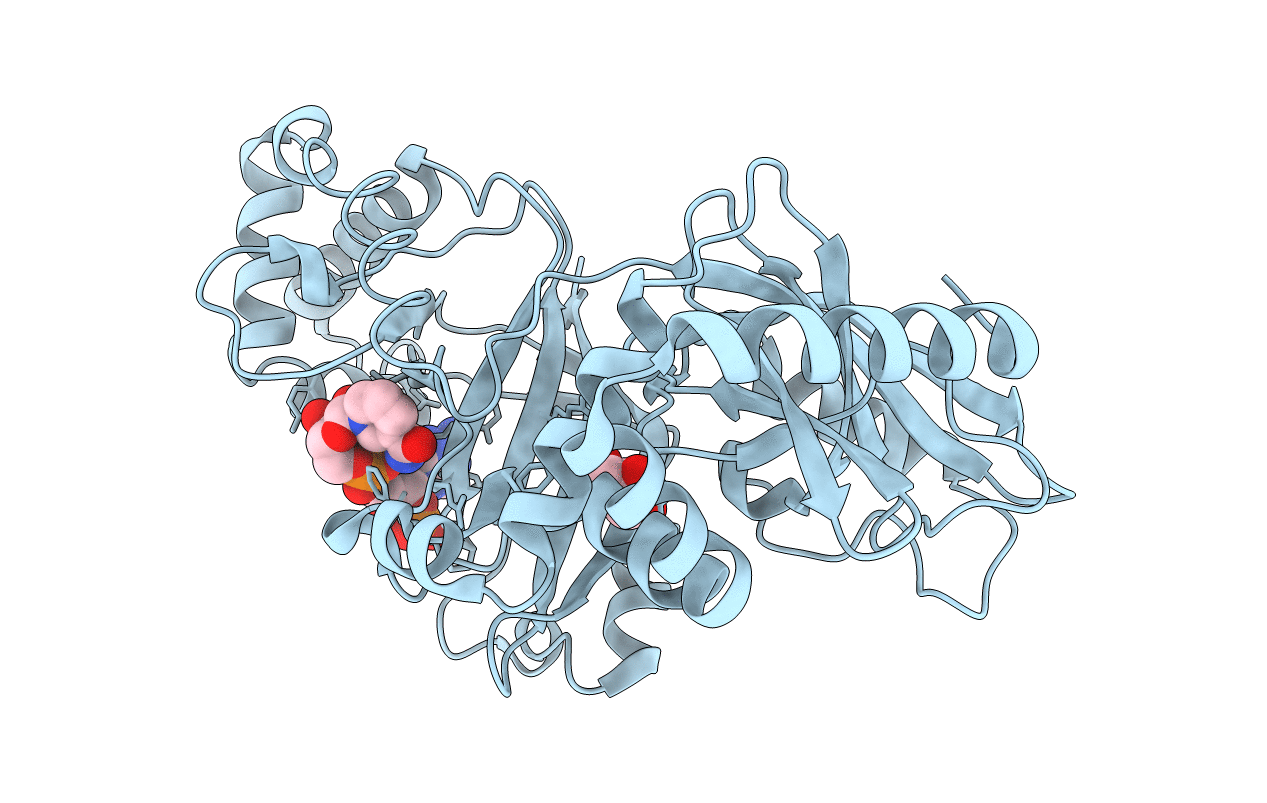 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2009-08-18 Classification: TRANSFERASE Ligands: NDP, GOL |
 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-08-18 Classification: TRANSFERASE Ligands: NAD, GOL |
 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-18 Classification: TRANSFERASE |

