Search Count: 782
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: ARG, HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG, PGL |
 |
Crystal Structure Of A Allulose Transcriptional Regulator From Agrobacterium Fabrum
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSCRIPTION Ligands: A1EGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: A1ELW |
 |
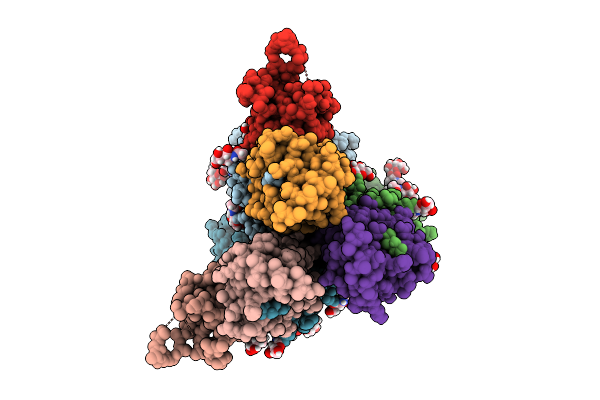
Cryoem Structure Of De Novo Antibody Fragment Scfv 6 With C. Difficile Toxin B (Tcdb)
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN |
 |
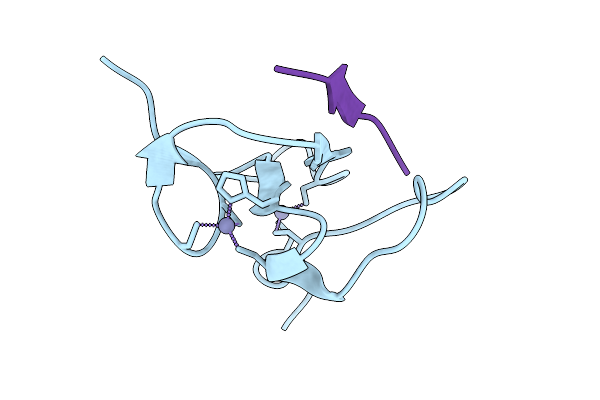
Cryoem Structure Of De Novo Vhh, Vhh_Flu_01, Bound To Influenza Ha, Strain A/Usa:Iowa/1943 H1N1.
Organism: Synthetic construct, Influenza a virus (strain a/usa:iowa/1943 h1n1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN Ligands: NAG |
 |
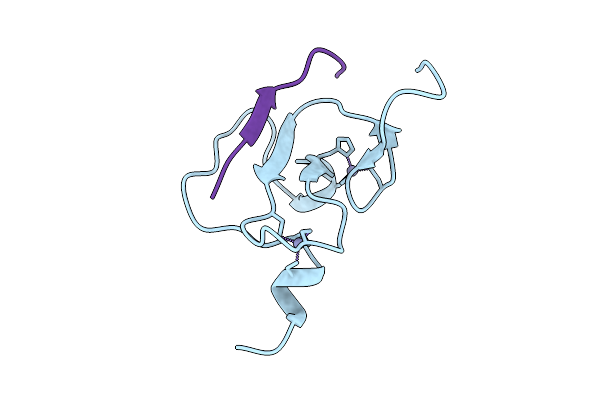
Crystal Structure Of Arabidopsis Thaliana Ing1 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
 |
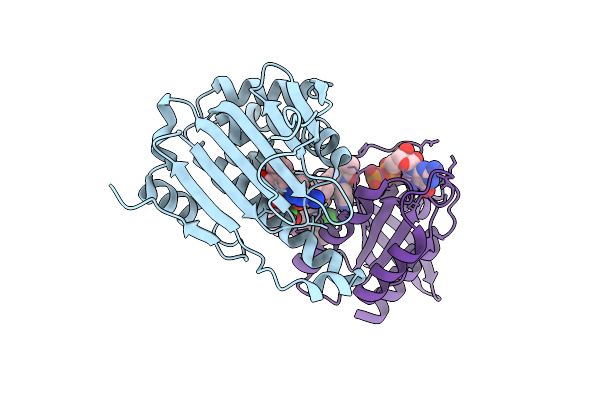
Crystal Structure Of Arabidopsis Thaliana Ing2 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: CHAPERONE/ONCOPROTEIN Ligands: BTB, CA, GDP, A1CRY, MG |
 |
Organism: Ruminococcus sp. xpd3002, Cloning vector pgrna-ccdb-1
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
 |
Organism: Ruminococcus sp. xpd3002, Cloning vector pgrna-ccdb-1
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
 |
Structure Of Fab Hb420 In Complex With Influenza H3N2 A/Moscow/10/1999 Neuraminidase
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Co-Crystal Structure Of Yeast Forkhead Transcription Factor Fkh1 Bound To Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: DNA BINDING PROTEIN/DNA Ligands: K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Apo Human Xpr1, Class 2, With Asymmetrically Dimerized Spx Domains
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: TRANSPORT PROTEIN |

