Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
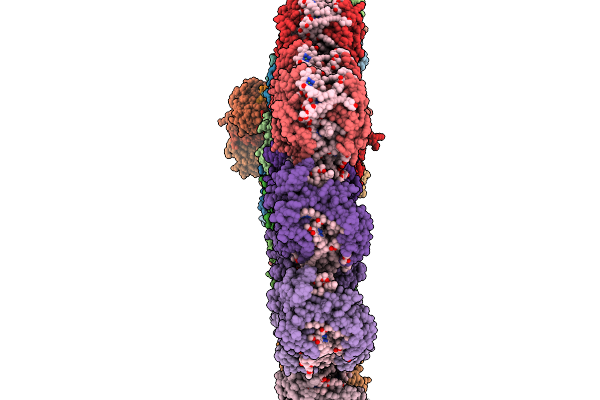 |
Organism: Emiliania huxleyi
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, DD6, LMG, A86, LHG, SQD, A1EB1, A1EB4, BCR, PQN, SF4, DGD |
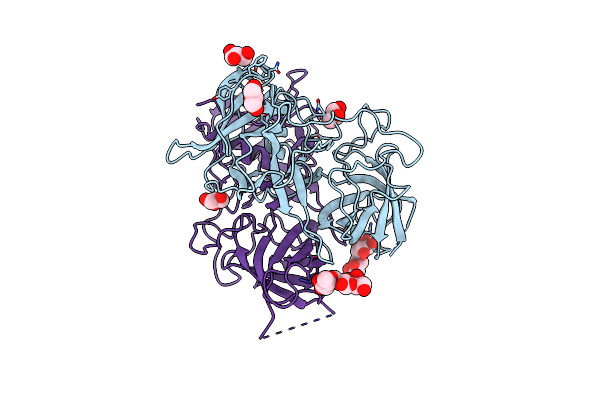 |
Functional And Structural Characterization Of Norovirus Gii.6 In Recognizing Histo-Blood Group Antigens
Organism: Norovirus gii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: GOL |
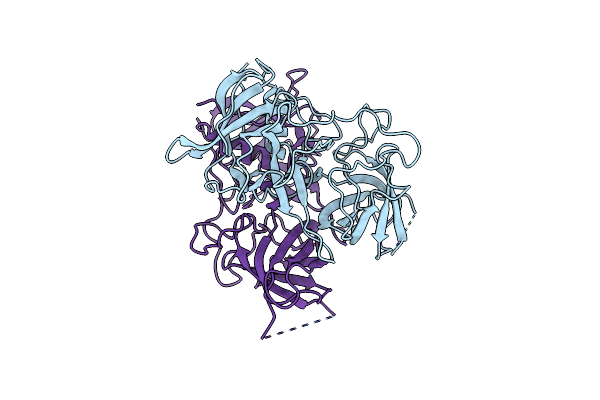 |
Functional And Structural Characterization Of Norovirus Gii.6 In Recognizing Histo-Blood Group Antigens
Organism: Norovirus gii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-03 Classification: VIRAL PROTEIN |
 |
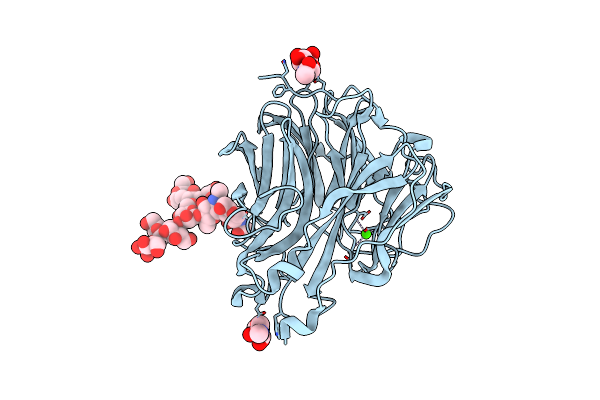
Gii.13/21 Noroviruses Recognize Glycans With A Terminal Beta-Galactose Via An Unconventional Glycan Binding Site
Organism: Human norovirus - alphatron
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-05-22 Classification: VIRAL PROTEIN |
 |
Gii.13/21 Noroviruses Recognize Glycans With A Terminal Beta-Galactose Via An Unconventional Glycan Binding Site
Organism: Human norovirus - alphatron
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-05-22 Classification: VIRAL PROTEIN |
 |
Gii.13/21 Noroviruses Recognize Glycans With A Terminal Beta-Galactose Via An Unconventional Glycan Binding Site
Organism: Human norovirus - alphatron
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-05-22 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Gii.13/21 Noroviruses Recognize Glycans With A Terminal Beta-Galactose Via An Unconventional Glycan Binding Site
Organism: Human norovirus - alphatron
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-05-22 Classification: VIRAL PROTEIN Ligands: THR |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, G39 |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, ZMR |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, LNV |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, BCZ |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, G39 |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, ZMR |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, LNV |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: NAG, CA, BCZ |
 |
Structural And Functional Characterization Of Neuraminidase-Like Molecule N10 Derived From Bat Influenza A Virus
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-09-19 Classification: HYDROLASE Ligands: CA, ZN, NAG |