Search Count: 269
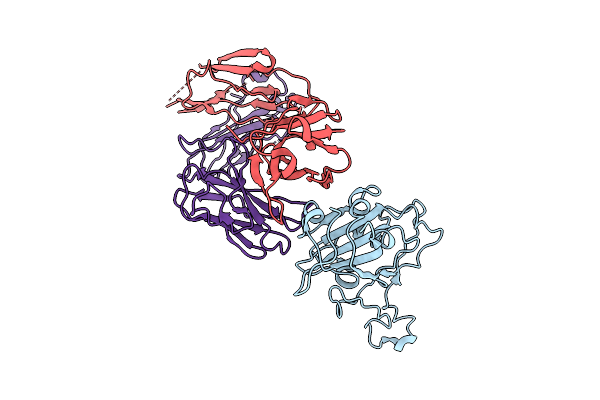 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
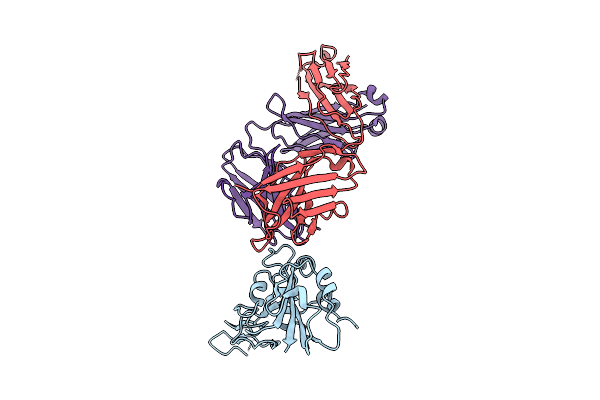
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
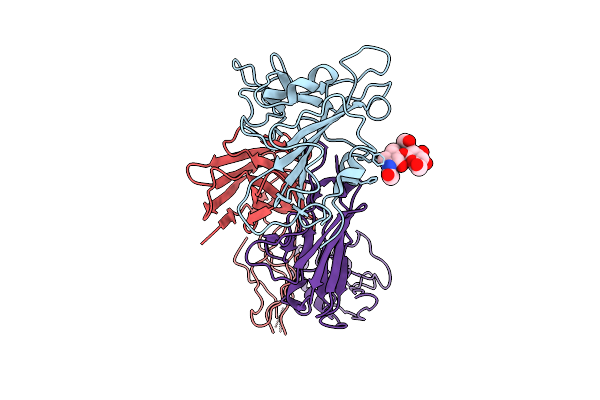
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
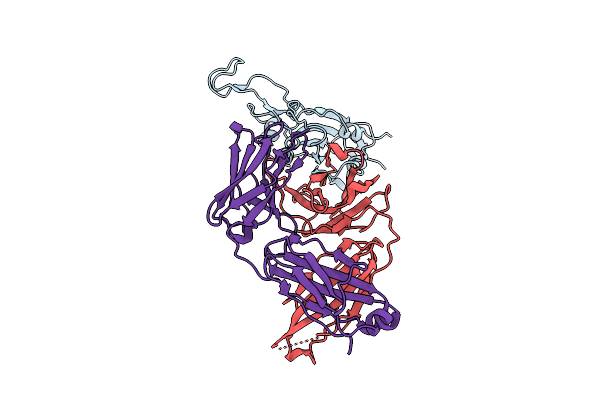
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
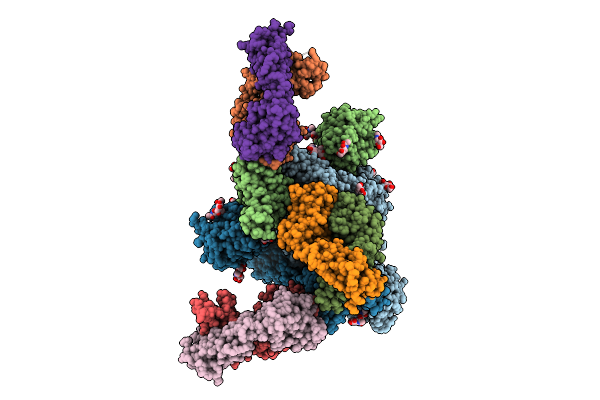
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
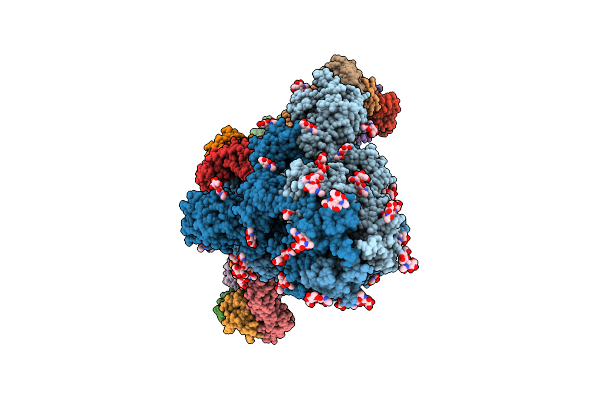
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
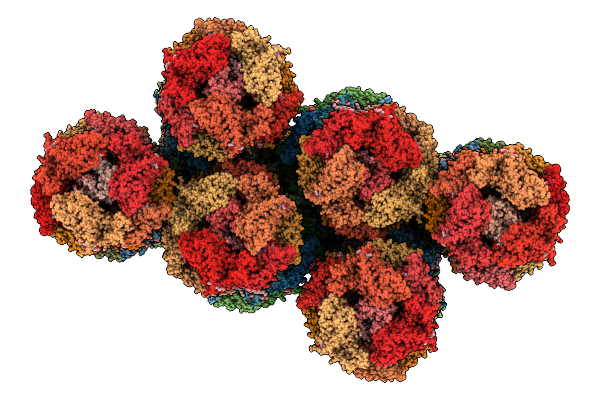
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
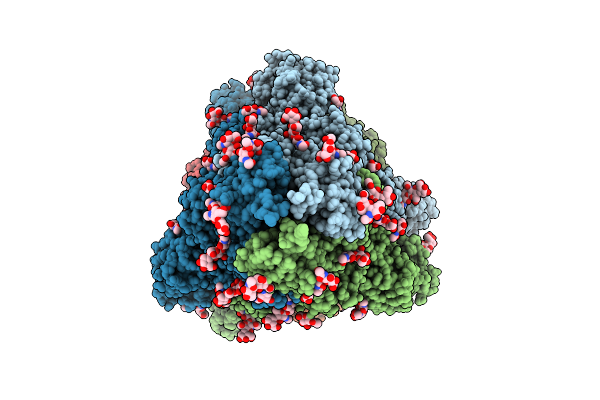
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
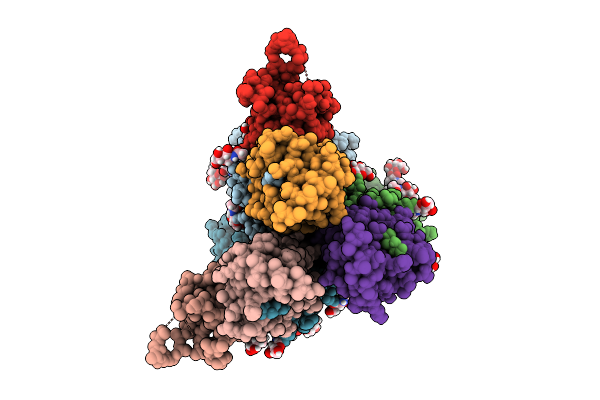
Cryoem Structure Of De Novo Antibody Fragment Scfv 6 With C. Difficile Toxin B (Tcdb)
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN |
 |
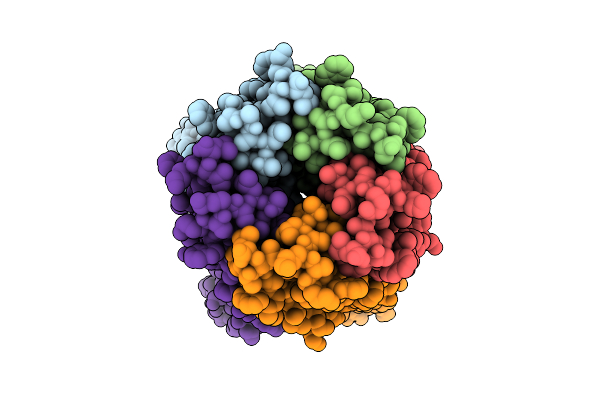
Cryoem Structure Of De Novo Vhh, Vhh_Flu_01, Bound To Influenza Ha, Strain A/Usa:Iowa/1943 H1N1.
Organism: Synthetic construct, Influenza a virus (strain a/usa:iowa/1943 h1n1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN Ligands: NAG |
 |
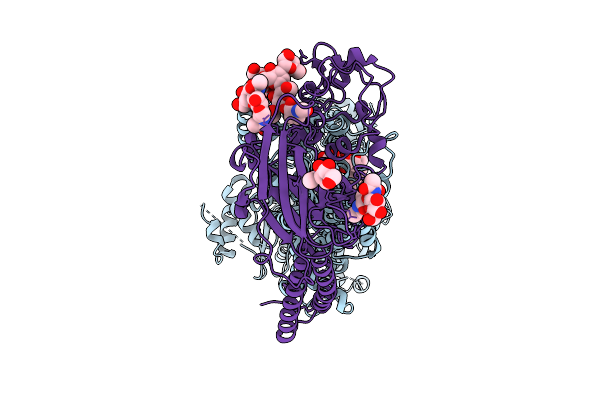
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DE NOVO PROTEIN Ligands: IOD |
 |
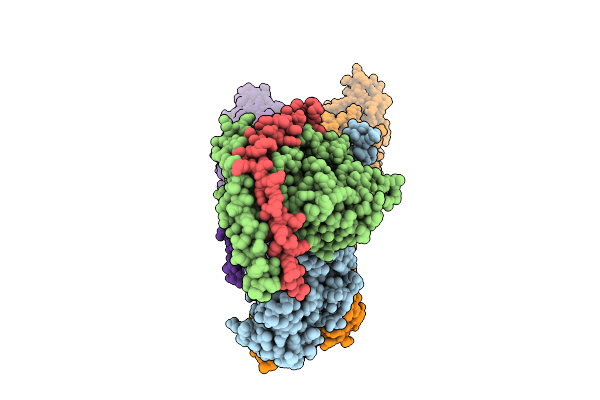
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: HVG, A1ENS |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: GLU |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: Z99 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: A1ENR, HVG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN Ligands: LMT |
 |
Cryo-Em Structure Of A De Novo Designed Voltage-Gated Anion Channel (Dvgac)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE/INHIBITOR Ligands: 8X7, ZN, ACT |

