Search Count: 460
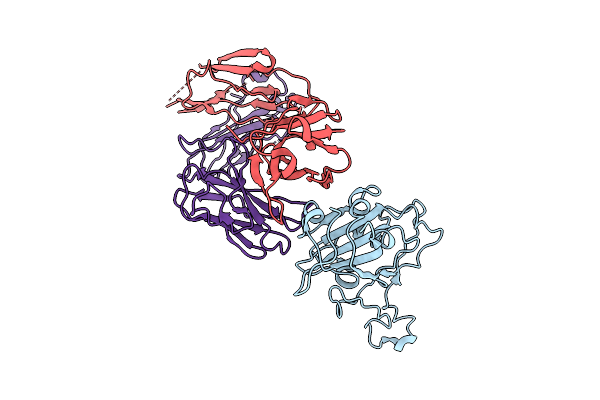 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
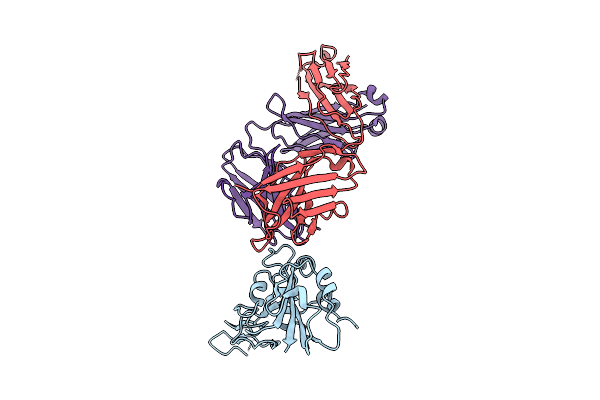
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
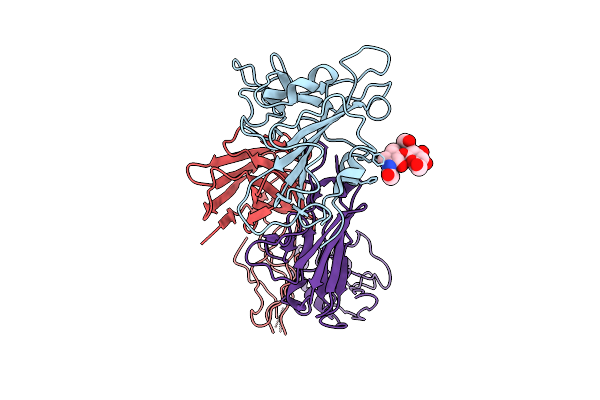
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
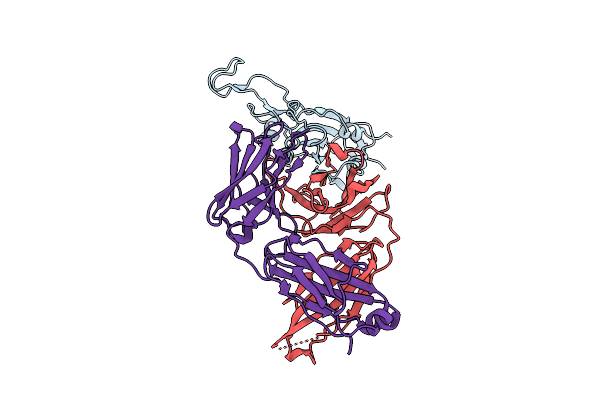
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
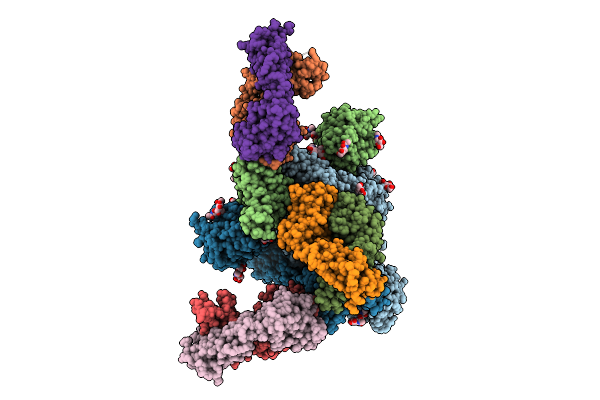
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
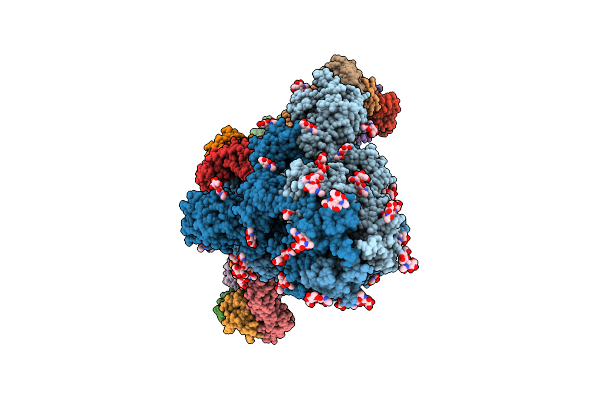
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
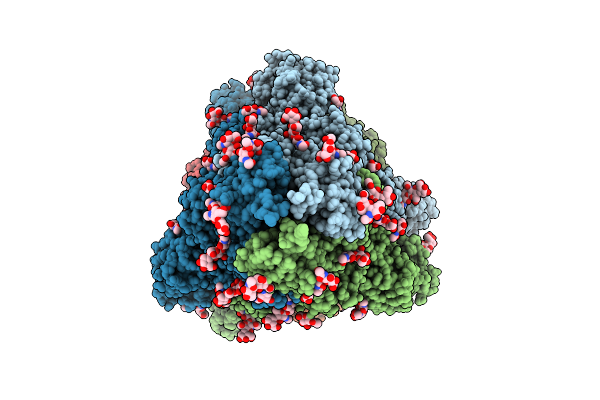
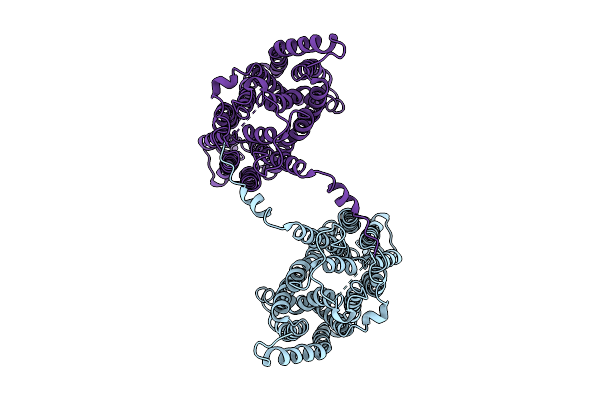
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
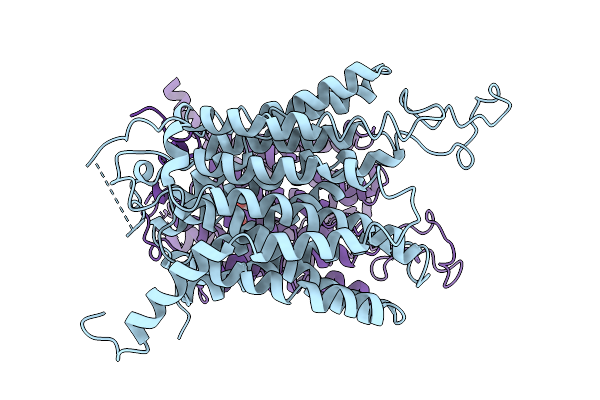
Cryo-Em Structure Of The Human G6P Transporter Slc37A2 In The Apo State With A Symmetric Er Luminal-Open Conformation.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN |
 |
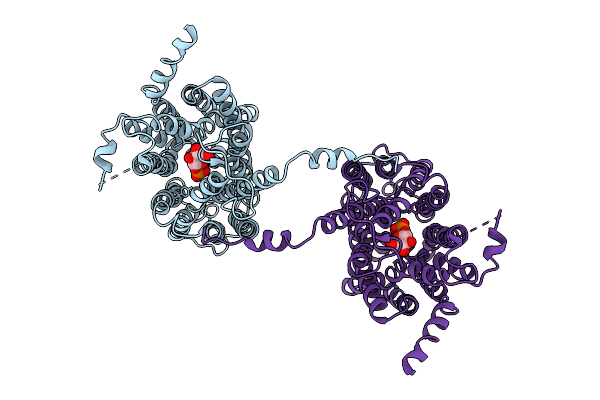
Cryo-Em Structure Of The Human G6P Transporter Slc37A2 In The Presence Of G6P With An Asymmetric Conformation.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: G6P |
 |
Cryo-Em Structure Of The Human G6P Transporter Slc37A2 In The G6P Bound State With A Symmetric Cytosolic-Open Conformation.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: G6P |
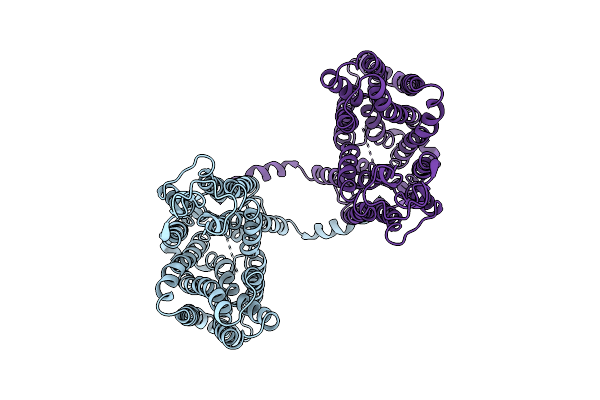 |
Cryo-Em Structure Of The Human G6P Transporter Slc37A2 In The Presence Of G6P With A Symmetric Er Luminal-Open Conformation.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN |
 |
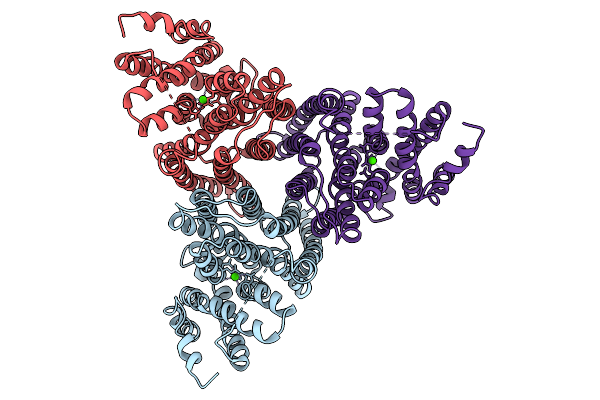
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Phosphomimetic Mutant Cax1 In Arabidopsis Thaliana In The Calcium-Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Structure Of The Phosphomimetic Mutant Cax1 In Arabidopsis Thaliana In The Apo State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Human Xpr1 In Presence Of Inorganic Phosphate And Phytic Acid
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Auxin Importer Aux1 In Arabidopsis Thaliana In The Iaa-Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN Ligands: IAC |
 |
Structure Of The Auxin Importer Aux1 In Arabidopsis Thaliana In The Apo State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Auxin Importer Aux1 In Arabidopsis Thaliana In The Chpaa-Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN Ligands: 3C4 |

