Search Count: 674
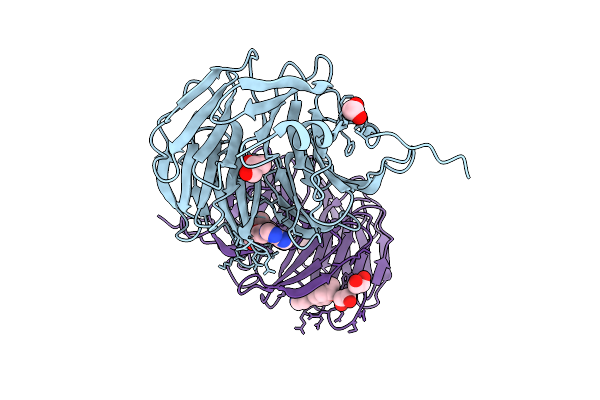 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: NUCLEAR PROTEIN Ligands: GOL, A1BI5, EDO |
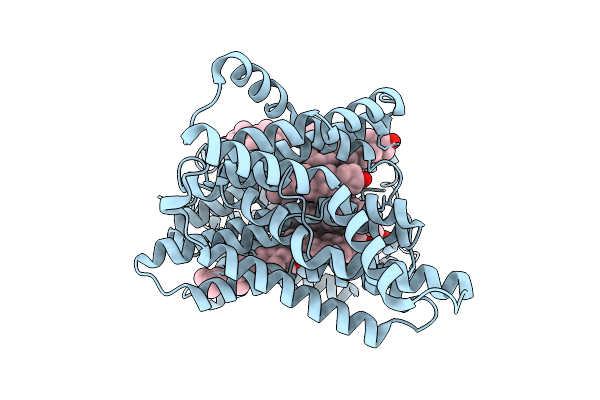 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: CLR |
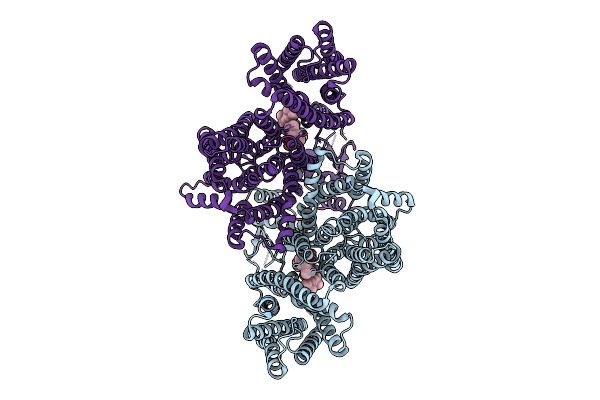 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: Y01, CLR |
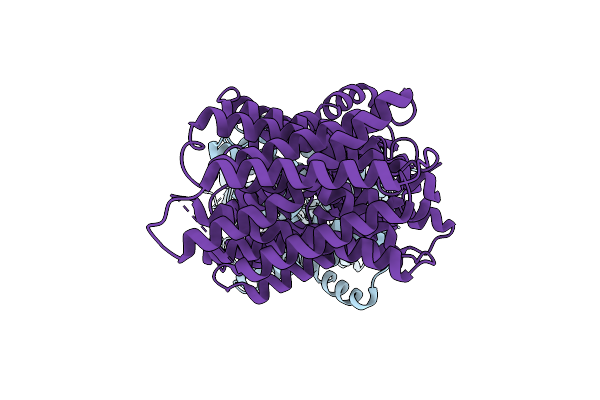 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak1
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak2
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Ectopseudomonas mendocina ymp
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: TRS, GOL, CA, EDO |
 |
Organism: Wuhan spiny eel influenza virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, NAG, A1JCF, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, NAG, A1JCG, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, A1JCH, PO4 |
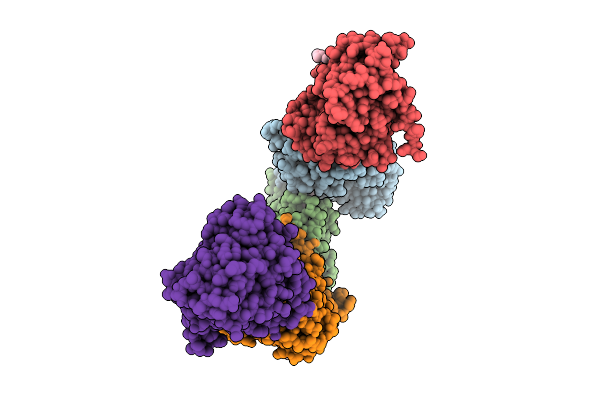 |
Crystal Structure Of Egfr Exon20 Insertion Mutant In Complex With Enozertinib (Oric-114)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SIGNALING PROTEIN Ligands: A1L8T |
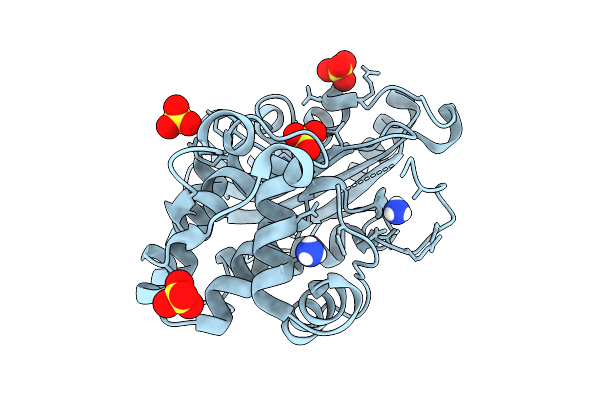 |
Structure Of P167S/D240G/D172A/S104G Blac From Mycobacterium Tuberculosis At Ph 5.5
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: NH4, SO4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: CIT |
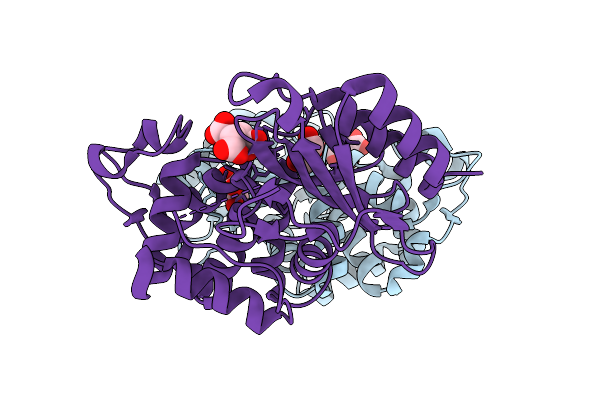 |
Structure Of P167S/D240G/D172A/S104G/H184R Blac From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: FLC, GOL |
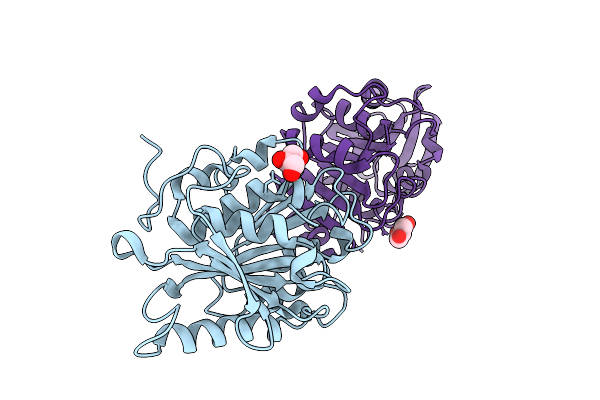 |
Organism: Pseudomonadota bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: GOL |
 |
 |
Cryo-Em Structure Of Mjhku4R-Cov-1 Receptor-Binding Domain Complexed With Human Cd26
Organism: Homo sapiens, Tylonycteris bat coronavirus hku4
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: CA, A1BBG |

