Search Count: 312
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, NAG, A1JCF, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, NAG, A1JCG, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, A1JCH, PO4 |
 |
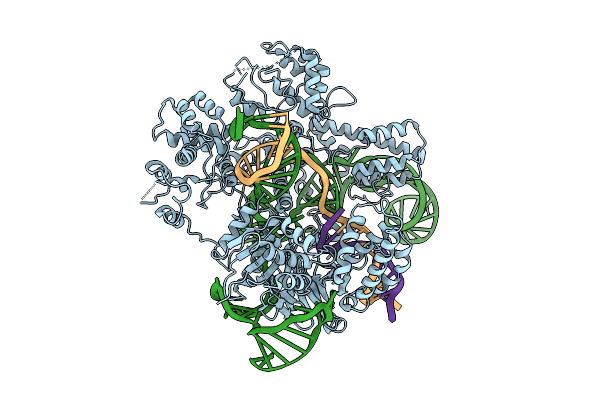
Cryo-Em Structure Of Frcas9 In Complex With Sgrna And 26-Nt Ts And 4-Nt Nts Substrates
Organism: Faecalibaculum rodentium, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: HYDROLASE/RNA/DNA Ligands: HOH |
 |
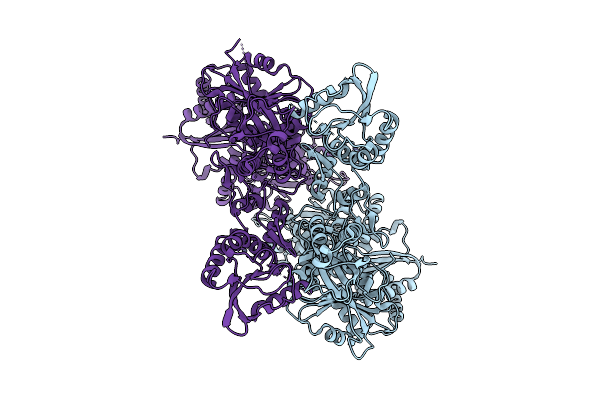
Cryo-Em Structure Of Frcas9 In Complex With Sgrna And 43-Bp Dsdna Substrate
Organism: Homo sapiens, Faecalibaculum rodentium
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: DNA/RNA/HYDROLASE |
 |
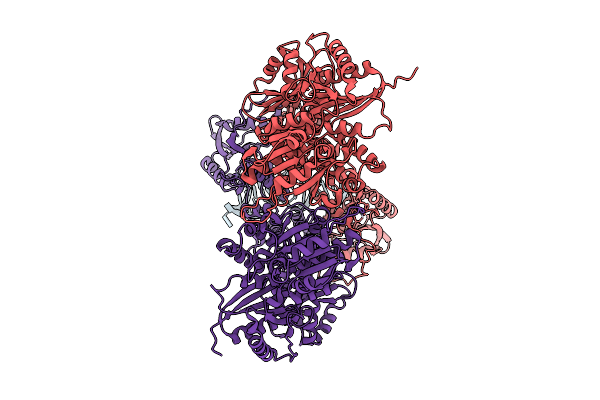
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
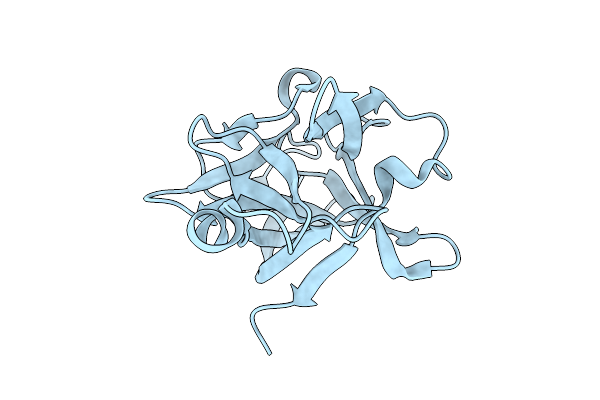
Electronic Microscopy Structure Of Human Schlafen14-E211A Dimer In Complex With Dsrna
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
 |
 |
 |
Pvtx-405: A Potent, Highly Selective, And Orally Efficacious Molecular Glue Degrader Of Ikzf2 For Cancer Immunotherapy
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-05-14 Classification: TRANSCRIPTION Ligands: A1A8N, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-05-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: XQF |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-04-23 Classification: VIRAL PROTEIN Ligands: XDQ, EDO, CL |
 |
 |
Cryo-Em Structure Of Neurotensin Receptor 1 In Complex With Beta-Arrestin1 And Sbi-553 (Complex 1)
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: SRW, PIO |
 |
Cryo-Em Structure Of Neurotensin Receptor 1 In Complex With Beta-Arrestin1 And Sbi-553 (Complex 3)
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: PIO, SRW |
 |
Cryo-Em Structure Of Neurotensin Receptor 1 In Complex With Beta-Arrestin1 And Sbi-553 (Complex 2)
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: PIO, SRW |
 |
Cryo-Em Structure Of Neurotensin Receptor 1 In Complex With Beta-Arrestin 1
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.41 Å Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: POV |
 |
Cryo-Em Structure Of Human Trpv3 In Complex With Citronellal Determined In Msp2N2 Nanodisc
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: POV, A1EBV |
 |
Cryo-Em Structure Of Human Trpv3 In Complex With Citral Determined In Msp2N2 Nanodisc
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: POV, GRQ |

