Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Group 1 Oligosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaca From Cohnella Abietis, Apo 1 Form
Organism: Cohnella abietis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Group 1 Oligosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaca From Cohnella Abietis, Apo 2 Form
Organism: Cohnella abietis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Group 2Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylhexosaminidase Ngaat From Arabidopsis Thaliana In Complex With Galnac-Thiazoline
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: GNL, PEG, CL |
 |
Crystal Structure Of Group 2 Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylhexosaminidase Ngaat From Arabidopsis Thaliana In Complex With Glcnac-Thiazoline
Organism: Arabidopsis thaliana x arabidopsis lyrata
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: PEG, NGT, CL |
 |
Crystal Structure Of Group 3 Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngadssm, Apo Form
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of Group 3 Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngadssm In Complex With Galnac-Thiazoline
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: GNL, GOL, SO4, CL |
 |
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngap2 From Paenibacillus Sp. Ts12, Apo Form
Organism: Paenibacillus sp. ts12
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: BR |
 |
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngap2 From Paenibacillus Sp. Ts12 In Complex With Galnac-Thiazoline
Organism: Paenibacillus sp. ts12
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: GNL, BR |
 |
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaly From Lacticaseibacillus Yichunensis, Apo Form
Organism: Lacticaseibacillus yichunensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-24 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: 4VZ, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: 4WD |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-06-26 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
A New Conserved Neutralizing Epitope At The Globular Head Of Hemagglutinin In H3N2 Influenza Viruses
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2014-04-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of The Catalytic Domain Of Pyrrolysyl-Trna Synthetase In Complex With Amppnp (Re-Refined)
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-01-02 Classification: LIGASE Ligands: ANP, MG |
 |
Crystal Structure Of The Semet Substituted Catalytic Domain Of Pyrrolysyl-Trna Synthetase
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-01-02 Classification: LIGASE Ligands: ANP, MG |
 |
Crystal Structure Of The Catalytic Domain Of Pyrrolysyl-Trna Synthetase In Triclinic Crystal Form
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-01-02 Classification: LIGASE Ligands: AMP, PO4, BLK |
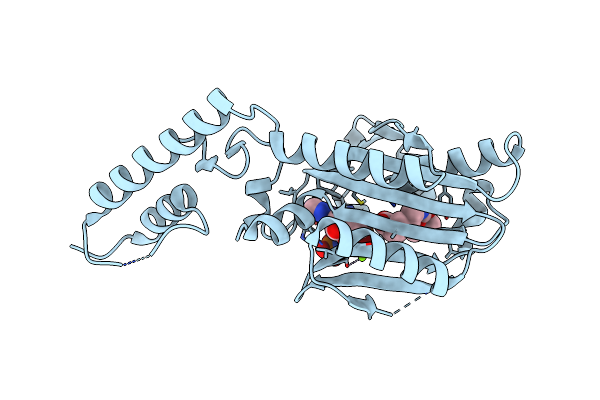 |
Crystal Structure Of The Catalytic Domain Of Pyrrolysyl-Trna Synthetase In Complex With Boclys And Amppnp (Form 2)
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-01-02 Classification: LIGASE Ligands: ANP, LBY, MG |
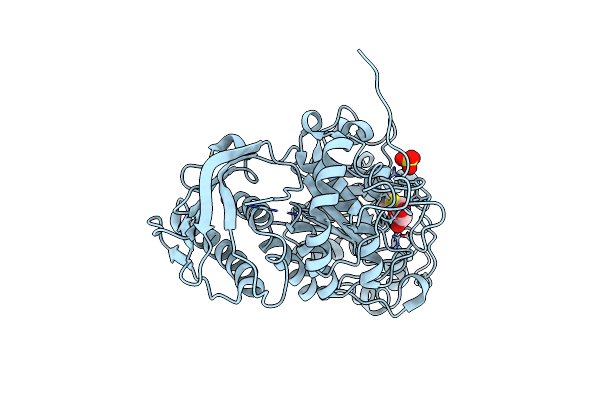 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Nag-Thiazoline.
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, NGT |
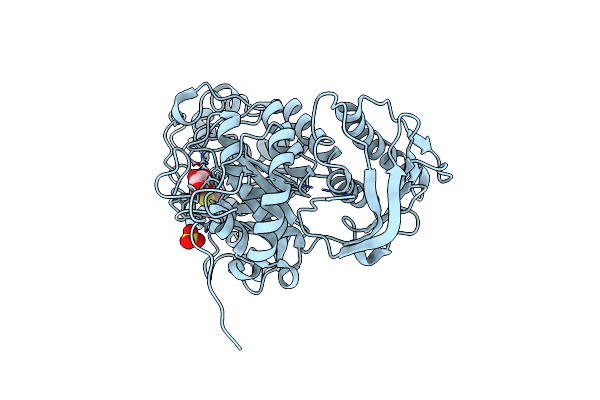 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Gal-Nag-Thiazoline
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, GNL |
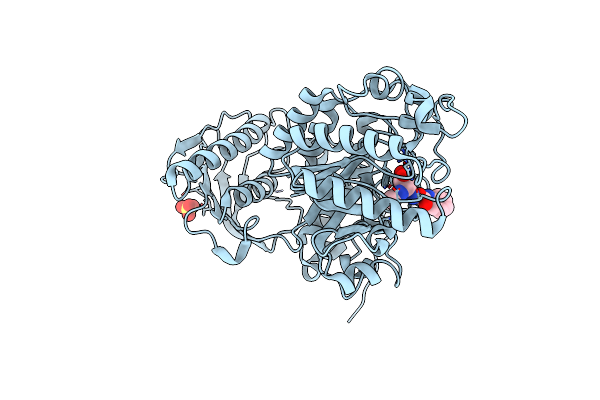 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Pugnac
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, OAN |