Search Count: 16
 |
Crystal Structure Of Neutralizing Antibody 80 In Complex With Sars-Cov-2 Receptor Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2023-05-24 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-02 Classification: CELL ADHESION Ligands: CL |
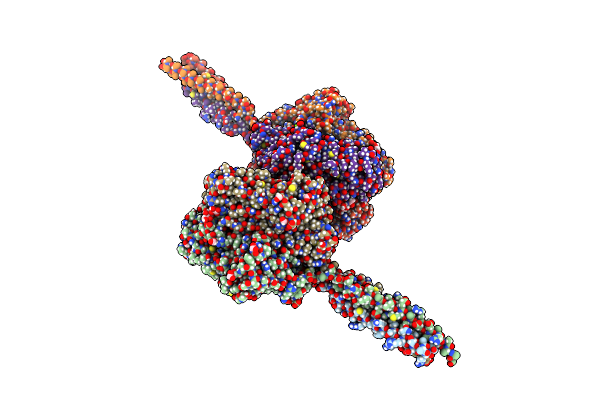 |
Crystal Structure Of The Infectious Salmon Anemia Virus (Isav) He Viral Receptor Complex
Organism: Infectious salmon anemia virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-22 Classification: VIRAL PROTEIN Ligands: MG, 79J, GOL, FMT |
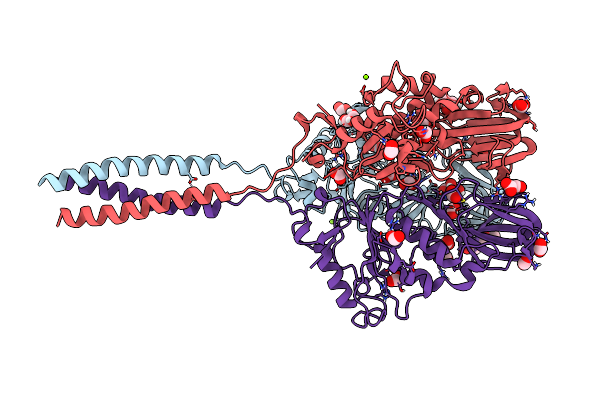 |
Crystal Structure Of The Infectious Salmon Anemia Virus (Isav) Hemagglutinin-Esterase Protein
Organism: Infectious salmon anemia virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-22 Classification: VIRAL PROTEIN Ligands: MG, GOL, FMT, 15P |
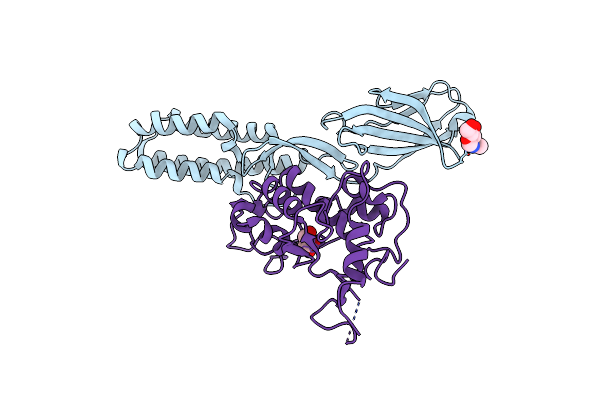 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-06-15 Classification: CELL ADHESION Ligands: NAG, CL, GOL |
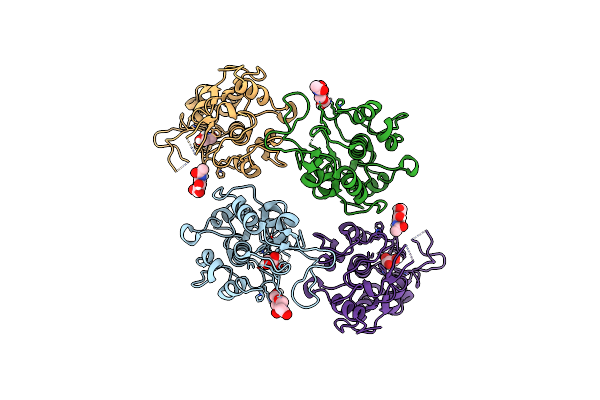 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-06-15 Classification: CELL ADHESION Ligands: NAG, CL, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2016-06-15 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-06-15 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-11-06 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-09-18 Classification: PROTEIN TRANSPORT Ligands: GNP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-26 Classification: PROTEIN TRANSPORT Ligands: GNP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-10-26 Classification: PROTEIN TRANSPORT Ligands: GDP, MG |
 |
Organism: Streptomyces galilaeus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2007-01-23 Classification: OXIDOREDUCTASE Ligands: AKY, FAD |
 |
Crystal Structure Of The Polyketide Cyclase Aknh With Bound Substrate And Product Analogue: Implications For Catalytic Mechanism And Product Stereoselectivity.
Organism: Streptomyces galilaeus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-02-14 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, NGV |
 |
Crystal Structure Of The Polyketide Cyclase Aknh With Bound Substrate And Product Analogue: Implications For Catalytic Mechanism And Product Stereoselectivity.
Organism: Streptomyces galilaeus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-02-14 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, AKV |
 |
Organism: Streptomyces nogalater
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2004-04-27 Classification: LYASE Ligands: NGV |

