Search Count: 30
 |
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSPORT PROTEIN Ligands: GTP, MG, GOL, NA, NO3 |
 |
Crystal Structure Of Tyrosine Phenol-Lyase In Complex With 3,5-Dihydroxybenzoic Acid
Organism: Morganella morganii subsp. morganii
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-04-23 Classification: LYASE Ligands: 34D, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2022-12-21 Classification: TRANSFERASE Ligands: ZN, SFG, J26, EDO, TRS |
 |
X-Ray Structure Of Apo-Vmfbpa, A Ferric Ion-Binding Protein From Vibrio Metschnikovii
Organism: Vibrio metschnikovii
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-12-15 Classification: METAL BINDING PROTEIN |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-23 Classification: TRANSPORT PROTEIN Ligands: GTP, MG, NO3, CL, EQF, GOL |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-06-09 Classification: TRANSPORT PROTEIN Ligands: MG, GTP, DMS, CL, G6U, GOL, MPO |
 |
Crystal Structure Of Dipeptidyl Peptidase 11 (Dpp11) With Citrate From Porphyromonas Gingivalis (Space)
Organism: Porphyromonas gingivalis (strain atcc 33277 / dsm 20709 / cip 103683 / jcm 12257 / nctc 11834 / 2561)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: PEG, CIT, GOL, K |
 |
Crystal Structure Of Dipeptidyl Peptidase 11 (Dpp11) With Sh-5 From Porphyromonas Gingivalis (Space)
Organism: Porphyromonas gingivalis (strain atcc 33277 / dsm 20709 / cip 103683 / jcm 12257 / nctc 11834 / 2561)
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: GOL, C8O |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae, Minute virus of mice
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-06-19 Classification: TRANSPORT PROTEIN Ligands: GTP, MG, EDO, GOL, NA |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Minute virus of mice
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-06-19 Classification: TRANSPORT PROTEIN Ligands: GTP, MG, EDO, GOL, NA |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Minute virus of mice
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2019-06-19 Classification: TRANSPORT PROTEIN Ligands: GTP, MG, EDO, GOL, NA |
 |
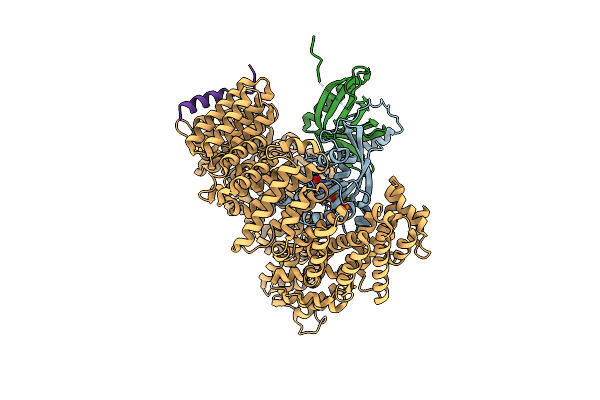
Organism: Homo sapiens, Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Minute virus of mice
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-06-19 Classification: TRANSPORT PROTEIN Ligands: GTP, MG, EDO, GOL, CL, NA |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Minute virus of mice
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-06-19 Classification: TRANSPORT PROTEIN Ligands: GTP, MG |
 |
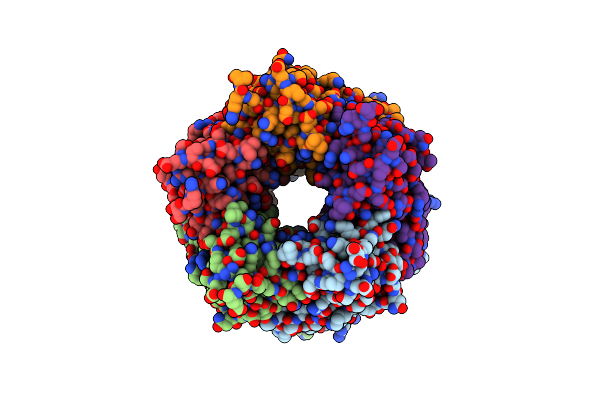
Crystal Structure Of Bace1 In Complex With (S)-N-(3-(2-Amino-6-(Fluoromethyl)-4 -Methyl-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5-Cyanopicolinamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-05-23 Classification: HYDROLASE/INHIBITOR Ligands: IOD, GOL, 0B6 |
 |
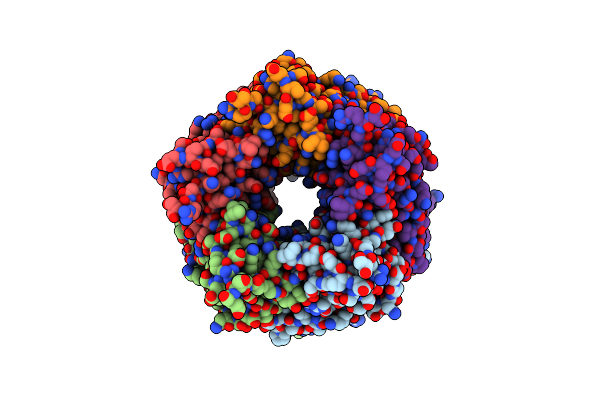
Crystal Structure Of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed With Imidacloprid
Organism: Lymnaea stagnalis
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2015-02-04 Classification: SIGNALING PROTEIN Ligands: IM4 |
 |
Crystal Structure Of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed With Clothianidin
Organism: Lymnaea stagnalis
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2015-02-04 Classification: SIGNALING PROTEIN Ligands: CT4 |
 |
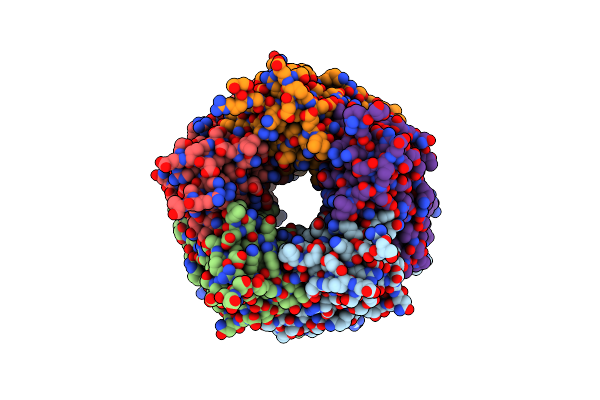
Crystal Structure Of Lymnaea Stagnalis Acetylcholine Binding Protein Complexed With Thiacloprid
Organism: Lymnaea stagnalis
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2015-02-04 Classification: SIGNALING PROTEIN Ligands: TH4 |
 |
Crystal Structure Of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed With Thiacloprid
Organism: Lymnaea stagnalis
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2015-02-04 Classification: SIGNALING PROTEIN Ligands: TH4 |
 |
Crystal Structure Of Lymnaea Stagnalis Acetylcholine Binding Protein Complexed With Nitromethylene Analogue Of Imidacloprid
Organism: Lymnaea stagnalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-02-04 Classification: SIGNALING PROTEIN Ligands: N1Y |
 |
Crystal Structure Of Lymnaea Stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed With Nitromethylene Analogue Of Imidacloprid
Organism: Lymnaea stagnalis
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2015-02-04 Classification: SIGNALING PROTEIN Ligands: N1Y |

