Search Count: 49
 |
Organism: Thalassiosira pseudonana
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: PLANT PROTEIN |
 |
Crystal Structure Of The Collagen Binding Domain Of Cnm From Streptococcus Mutans
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-04-10 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2020-11-11 Classification: IMMUNE SYSTEM Ligands: EWL, NAG, SO4 |
 |
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-11 Classification: IMMUNE SYSTEM Ligands: EWU, NAG, SO4 |
 |
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2020-11-11 Classification: IMMUNE SYSTEM Ligands: NAG, SO4, EWX |
 |
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-11 Classification: IMMUNE SYSTEM Ligands: NAG, SO4, EX0 |
 |
Organism: Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: IMMUNE SYSTEM Ligands: EX3 |
 |
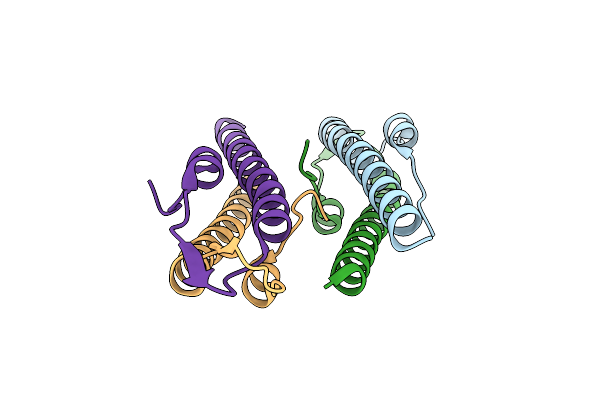
Cell Division Regulator, B. Subtilis Gpsb, In Complex With Peptide Fragment Of Penicillin Binding Protein Pbp1A
Organism: bacillus subtilis subsp. subtilis str. 168, Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-01-23 Classification: CELL CYCLE Ligands: MG |
 |
Cell Division Regulator Gpsb In Complex With Peptide Fragment Of L. Monocytogenes Penicillin Binding Protein Pbpa1
Organism: bacillus subtilis subsp. subtilis str. 168, Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-01-23 Classification: CELL CYCLE Ligands: ZN, IMD |
 |
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-23 Classification: CELL CYCLE |
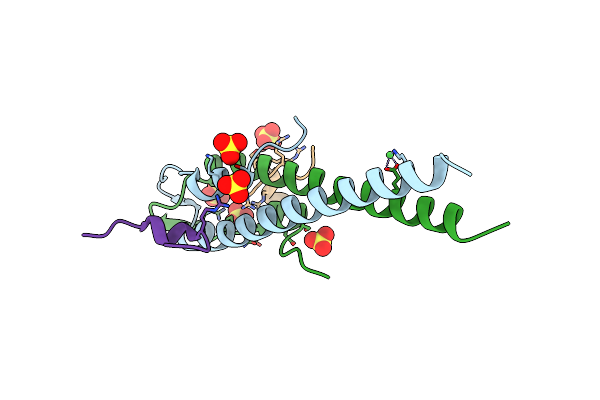 |
Cell Division Regulator, S. Pneumoniae Gpsb, In Complex With Peptide Fragment Of Penicillin Binding Protein Pbp2A
Organism: Streptococcus pneumoniae r6, Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-01-23 Classification: CELL CYCLE Ligands: SO4, NI |
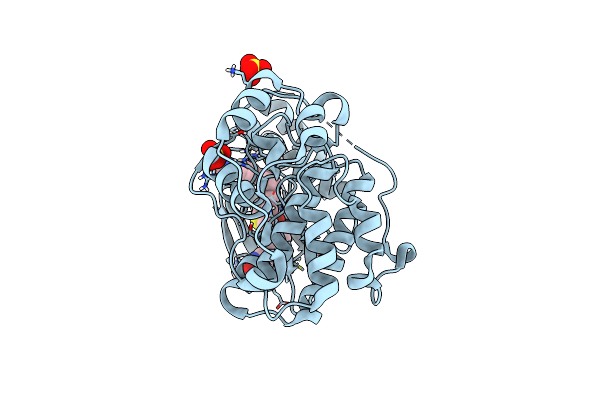 |
Discovery Of Mk-8353: An Orally Bioavailable Dual Mechanism Erk Inhibitor For Oncology
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-08-08 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, G67 |
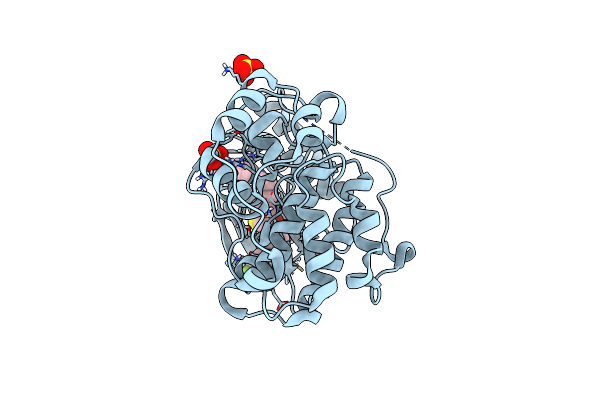 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-05-23 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: SO4, F8V |
 |
Organism: Palmaria palmata
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2016-10-05 Classification: PHOTOSYNTHESIS Ligands: CYC, PUB |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-08-17 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: 6YD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2016-08-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6YE |
 |
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, PRU |
 |
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, BGC |
 |
Crystal Structure Of Alpha-Glucosidase Mutant D202N In Complex With Glucose And Glycerol
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, BGC |
 |
Crystal Structure Of Alpha-Glucosidase Mutant E271Q In Complex With Maltose
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL |

