Search Count: 128
 |
Organism: Cucumis sativus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-03-08 Classification: SUGAR BINDING PROTEIN Ligands: NBZ, EDO |
 |
Organism: Cucumis sativus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-03-08 Classification: SUGAR BINDING PROTEIN Ligands: EDO |
 |
Organism: Cucumis sativus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-03-08 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Cucumis sativus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-03-08 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Cucumis sativus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-02-01 Classification: SUGAR BINDING PROTEIN Ligands: PT |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-03-09 Classification: SUGAR BINDING PROTEIN Ligands: MMA |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2022-03-09 Classification: SUGAR BINDING PROTEIN Ligands: GOL, EDO, PEG |
 |
Crystal Structure Of Retroviral Protease-Like Domain Of Ddi1 From Toxoplasma Gondii
Organism: Toxoplasma gondii gt1
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Retroviral Protease-Like Domain Of Ddi1 From Cryptosporidium Hominis
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-02-09 Classification: HYDROLASE |
 |
Structural And Functional Analysis Of Small Heat Shock Protein From Synechococcus Phage S-Shm2
Organism: Synechococcus phage s-shm2
Method: X-RAY DIFFRACTION Resolution:7.00 Å Release Date: 2021-07-21 Classification: CHAPERONE |
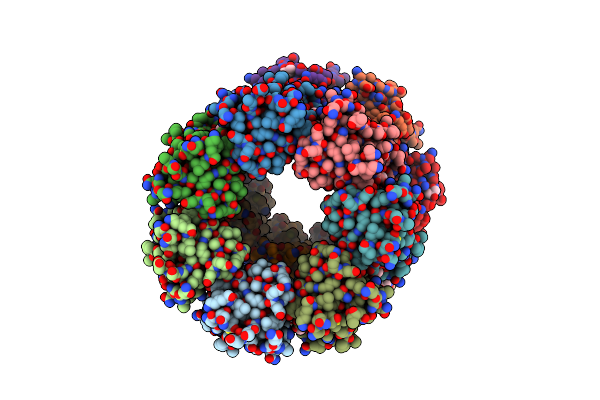 |
Organism: Methanococcus voltae (strain atcc baa-1334 / a3)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: GOL, EDO |
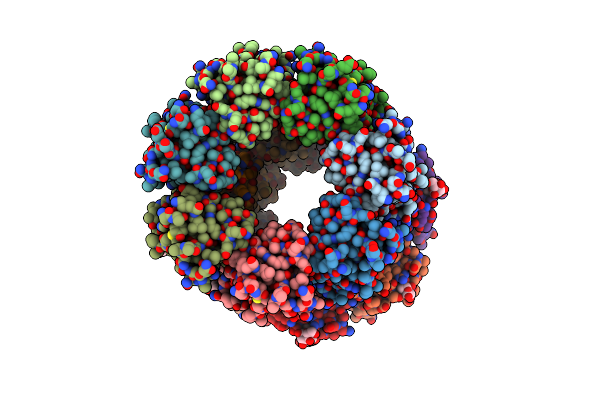 |
Organism: Methanococcus voltae (strain atcc baa-1334 / a3)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: MAN, ALA |
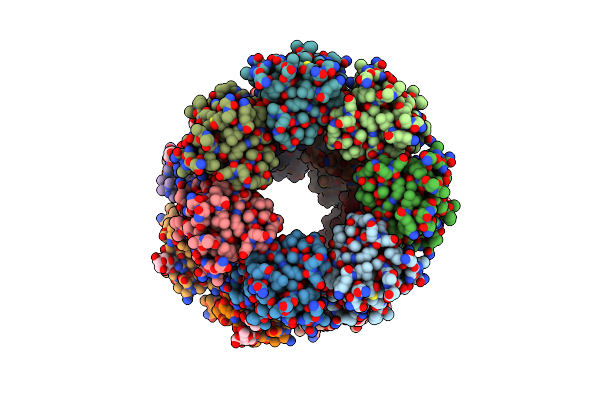 |
Organism: Methanococcus voltae (strain atcc baa-1334 / a3)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: MAN |
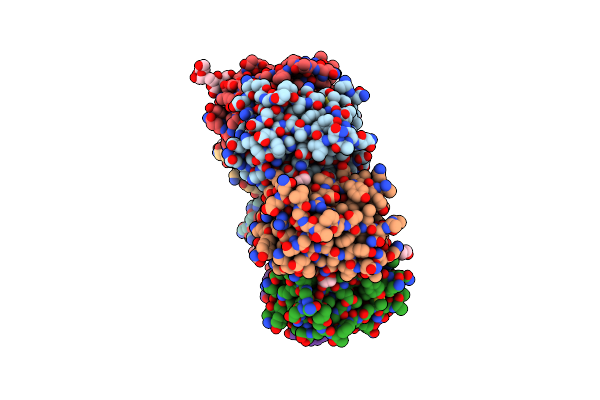 |
Organism: Methanococcus voltae (strain atcc baa-1334 / a3)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: EDO, GOL |
 |
Organism: Methanococcus voltae (strain atcc baa-1334 / a3)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: MAN |
 |
Organism: Methanococcus voltae (strain atcc baa-1334 / a3)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
Organism: Methanococcus voltae (strain atcc baa-1334 / a3)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: MAN, GOL |
 |
Purification, Characterization And X-Ray Structure Of Yhda-Type Azoreductase From Bacillus Velezensis
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2020-12-23 Classification: OXIDOREDUCTASE Ligands: GOL, PO4, FMN |
 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2020-11-04 Classification: CHAPERONE |
 |
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-25 Classification: SUGAR BINDING PROTEIN Ligands: GOL, IPA |

