Search Count: 116
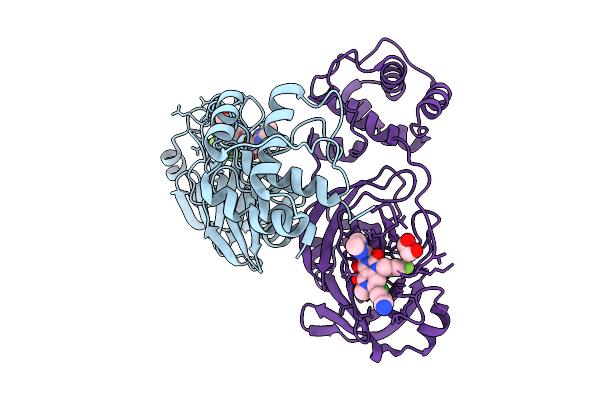 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: A1MA2, EDO |
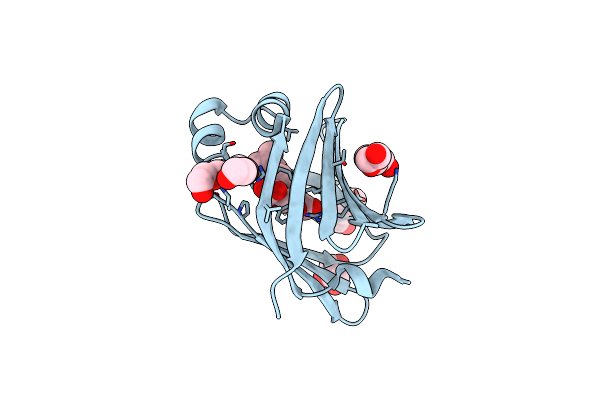 |
X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With R-Ibuprofen
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-04-30 Classification: LIPID BINDING PROTEIN Ligands: IZP, PG4, PEG, ACY, PGE, GOL |
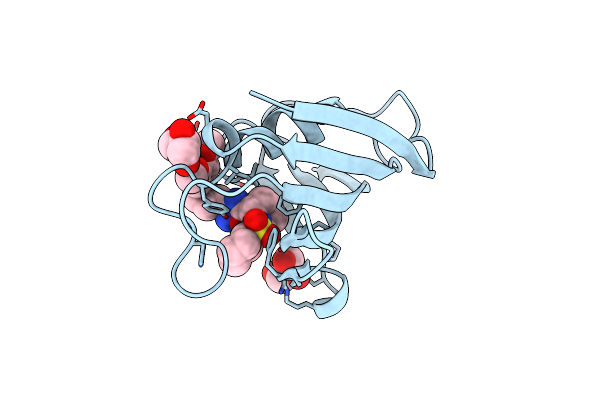 |
Crystal Structure Of Fkbp12 Complexed With Small Molecule Anchor For Protein-201
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2025-03-26 Classification: ISOMERASE Ligands: A1L7S, GOL |
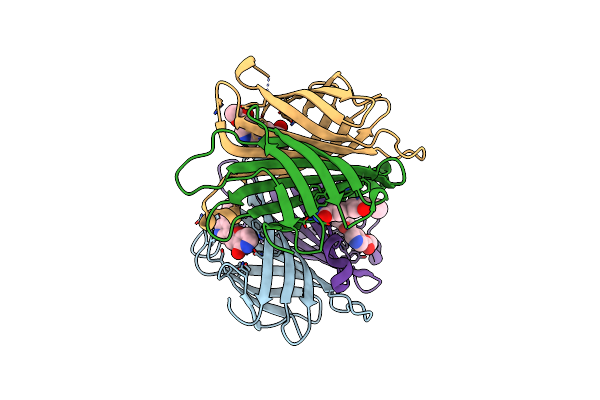 |
X-Ray Crystal Structure Of Alis4-Streptavidine Complex Without Cryo-Protectant Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: 6FX |
 |
X-Ray Crystal Structure Of Alis4-Streptavidine Complex With 10% Glycerol Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: DMS, GOL, PEG |
 |
X-Ray Crystal Structure Of Alis4-Streptavidine Complex With 20% Glycerol Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: GOL, DMS, PEG |
 |
X-Ray Crystal Structure Of Alis5-Streptavidine Complex Without Cryo-Protectant Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: DMS, A1LXQ |
 |
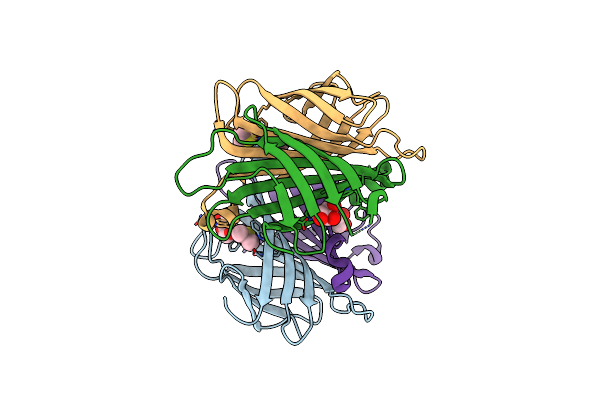
X-Ray Crystal Structure Of Alis5-Streptavidine Complex With 10% Glycerol Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: DMS, GOL, A1LXQ |
 |
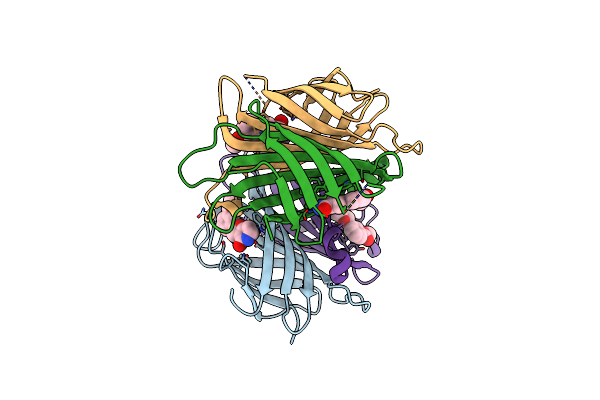
X-Ray Crystal Structure Of Alis5-Streptavidine Complex With 20% Glycerol Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: GOL, DMS |
 |
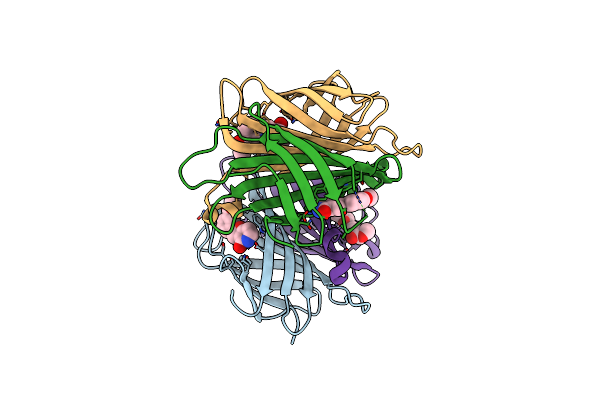
X-Ray Crystal Structure Of Alis2-Streptavidine Complex Without Cryo-Protectant Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: MT6 |
 |
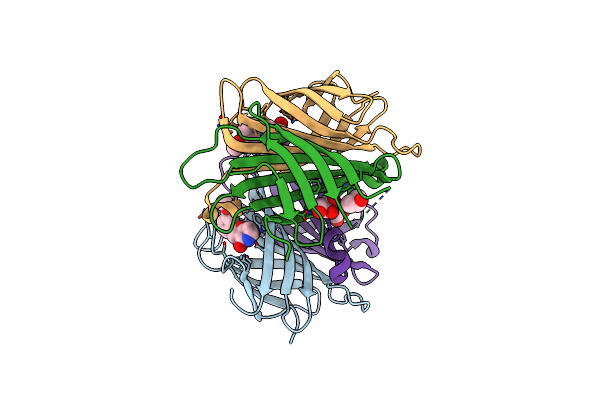
X-Ray Crystal Structure Of Alis2-Streptavidine Complex With 10% Glycerol Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: MT6 |
 |
X-Ray Crystal Structure Of Alis2-Streptavidine Complex With 20% Glycerol Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-29 Classification: CYTOSOLIC PROTEIN Ligands: MT6, GOL |
 |
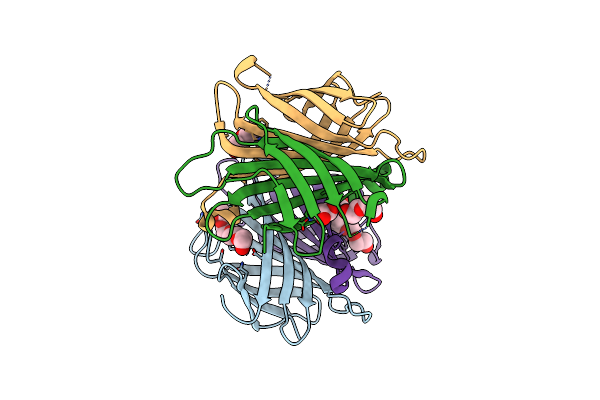
X-Ray Crystal Structure Of Streptavidin Flash-Cooled In 30% Glycerol At Ambient Pressure
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: PG6, PG4, GOL, PEG |
 |
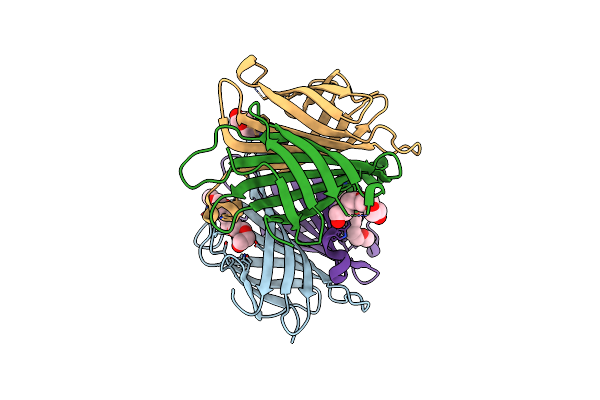
X-Ray Crystal Structure Of Streptavidin Flash-Cooled In 30% Peg1000 At Ambient Pressure
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: PG4, PEG |
 |
X-Ray Crystal Structure Of Streptavidine Without Cryo-Protectant Using A High-Pressure Cryocooling Method
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: PG6, PG4 |
 |
The 0.88 Angstrom X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Behenic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.88 Å Release Date: 2023-04-12 Classification: LIPID BINDING PROTEIN Ligands: EO3, P6G |
 |
The 0.88 Angstrom X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Tricosanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.88 Å Release Date: 2023-04-12 Classification: LIPID BINDING PROTEIN Ligands: F23, P6G, 1PE |
 |
The 0.92 Angstrom X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Lignoceric Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2023-03-29 Classification: LIPID BINDING PROTEIN Ligands: BZV, 1PE, GLP |
 |
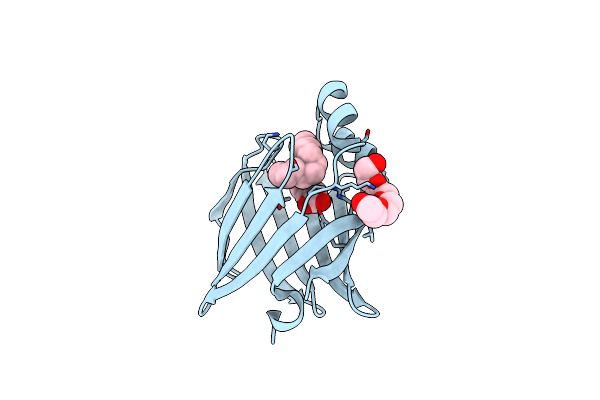
The 0.86 Angstrom X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Pelargonic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.86 Å Release Date: 2023-03-08 Classification: LIPID BINDING PROTEIN Ligands: KNA, P6G |
 |
The 0.96 Angstrom X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Arachidic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2023-03-08 Classification: LIPID BINDING PROTEIN Ligands: DCR, P6G |

