Search Count: 83
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: NUCLEAR PROTEIN/DNA |
 |
Crystal Structure Of A Highly Photostable And Bright Green Fluorescent Protein At Ph8.5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-10-04 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Crystal Structure Of A Highly Photostable And Bright Green Fluorescent Protein At Ph5.6
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-10-04 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-25 Classification: NUCLEAR PROTEIN/DNA Ligands: CL, MN |
 |
Crystal Structure Of Cystathionine Gamma-Lyase From Lactobacillus Plantarum Complexed With L-Serine
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-10-07 Classification: LYASE Ligands: KOU, PO4 |
 |
Crystal Structure Of Cystathionine Gamma-Lyase From Lactobacillus Plantarum Complexed With Cystathionine
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-10-07 Classification: LYASE Ligands: E9U, PO4 |
 |
Organism: Homo sapiens, Murine respirovirus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-19 Classification: VIRAL PROTEIN |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-12-18 Classification: LIGASE Ligands: TLA, ADP, MG |
 |
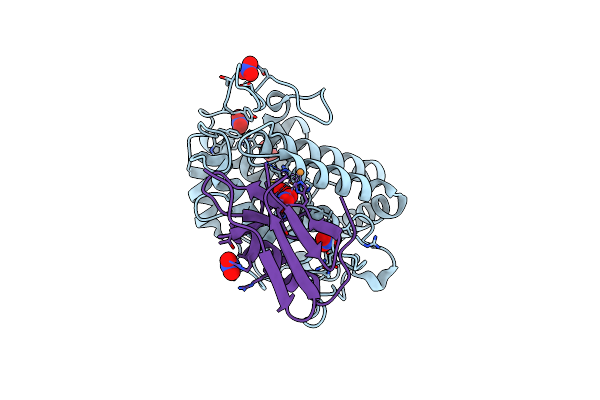
1.16 A-Resolution Crystal Structure Of The Deoxy-Form Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, NO3 |
 |
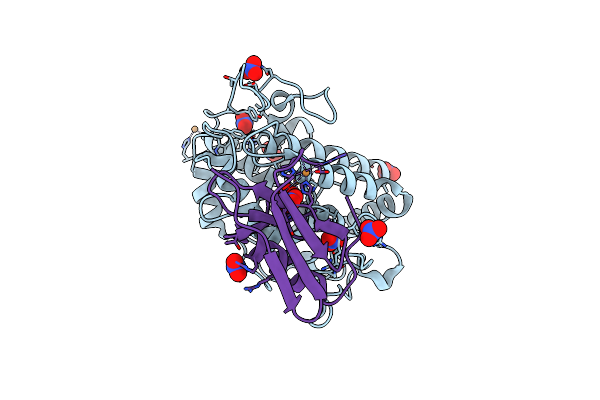
Crystal Structure Of Copper-Bound Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Y98F Mutant Of The Caddie Protein Obtained By Soaking In The Hydroxylamine-Containing Solution For 2 H At 298 K
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, PER, NO3 |
 |
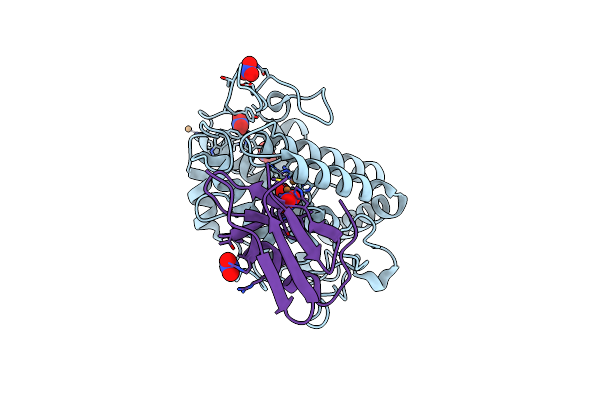
Crystal Structure Of Copper-Bound Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Hydroxylamine-Containing Solution For 10 Min At 298 K
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, PER, NO3 |
 |
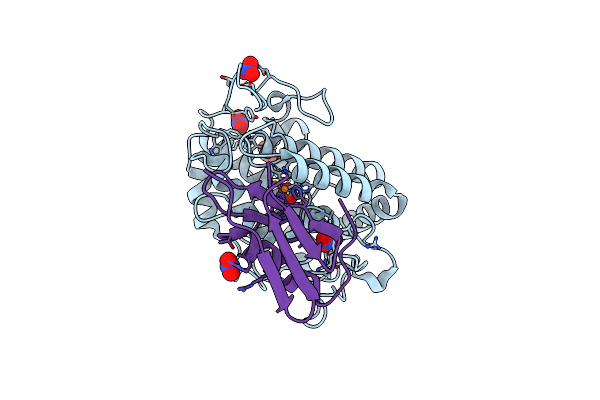
Crystal Structure Of Copper-Bound Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Hydroxylamine-Containing Solution For 20 Min At 298 K
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, PER, NO3 |
 |
Crystal Structure Of Copper-Bound Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Hydroxylamine-Containing Solution For 2 H At 298 K
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, PER, NO3 |
 |
Crystal Structure Of Copper-Bound Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Hydroxylamine-Containing Solution For 1 H At 277 K
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, PER, NO3 |
 |
Crystal Structure Of Copper-Bound Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Hydroxylamine-Containing Solution For 2 H At 277 K
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, PER, NO3 |
 |
Crystal Structure Of Copper-Bound Tyrosinase From Streptomyces Castaneoglobisporus In Complex With The Caddie Protein Obtained By Soaking In The Hydroxylamine-Containing Solution For 4 H At 277 K
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Release Date: 2018-12-26 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: CU, PER, NO3 |

