Search Count: 20
 |
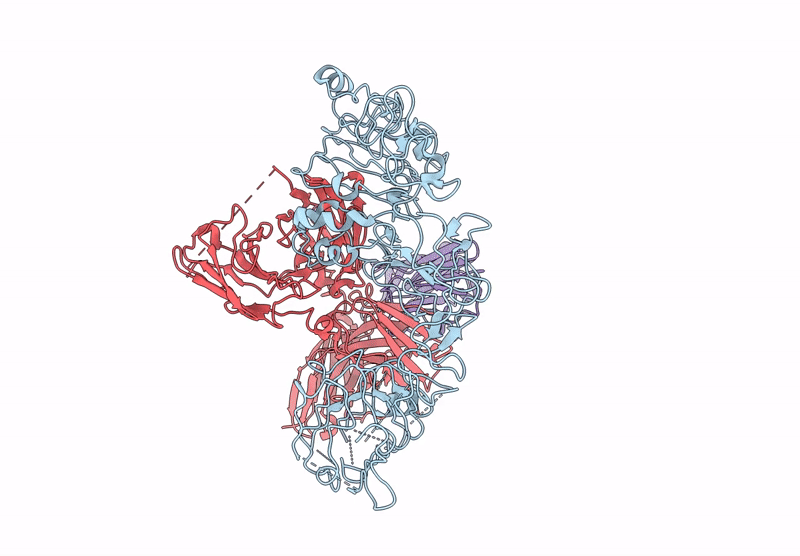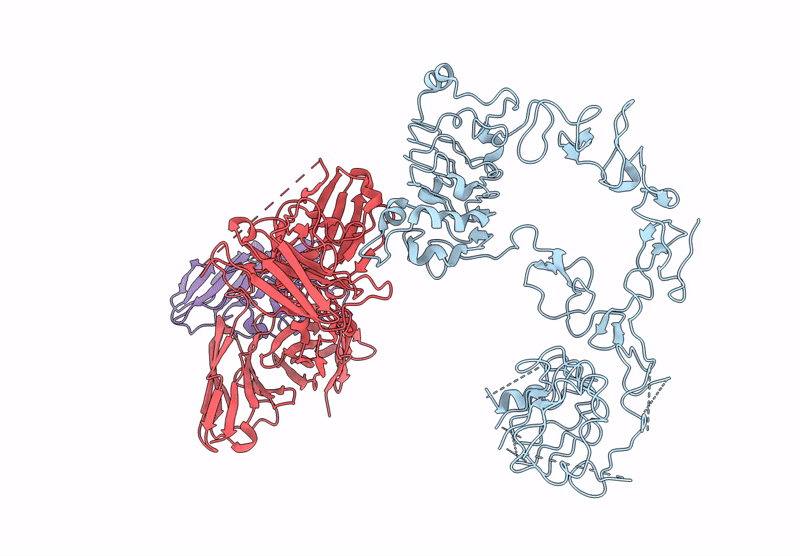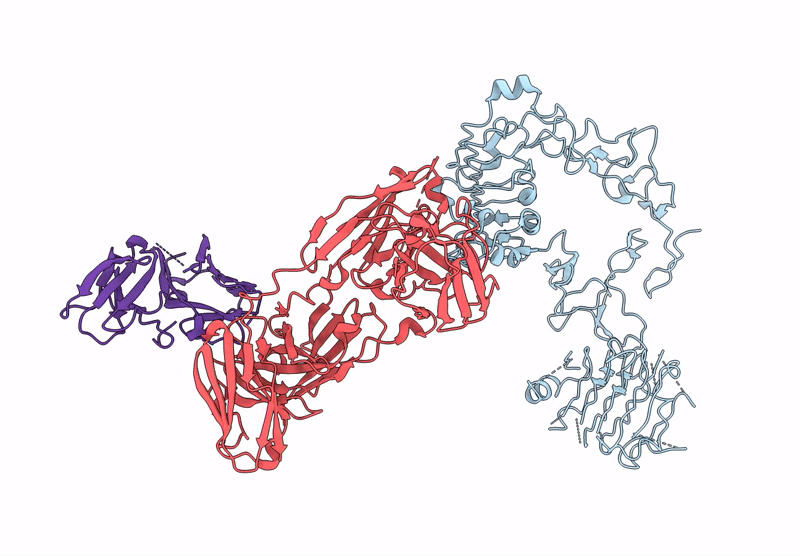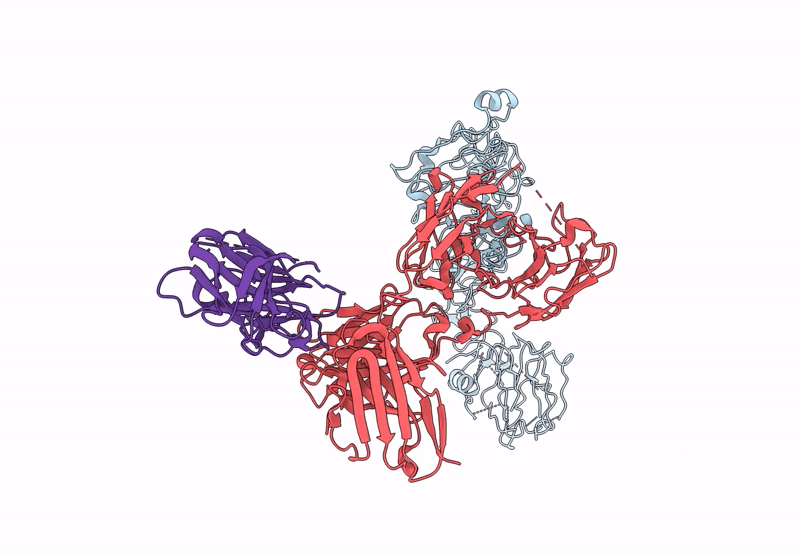
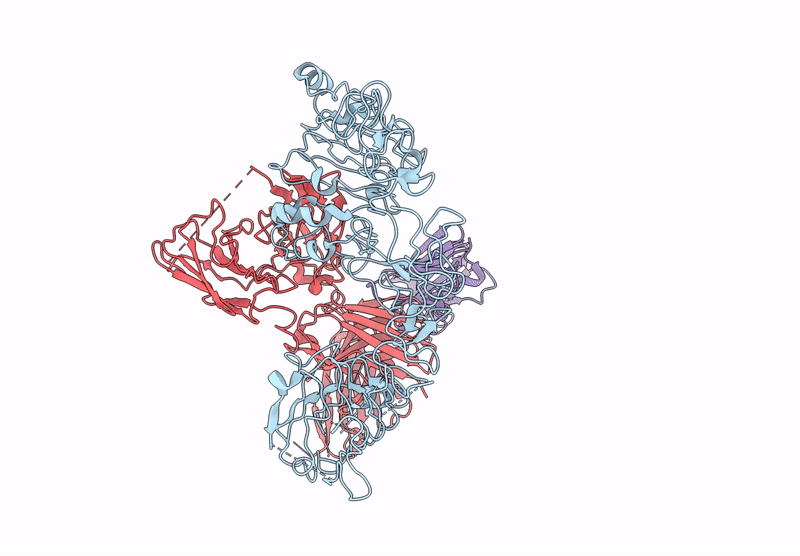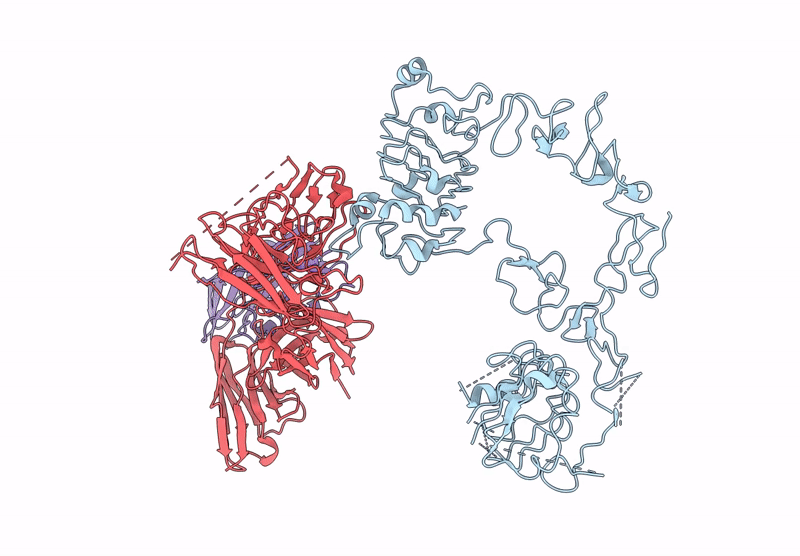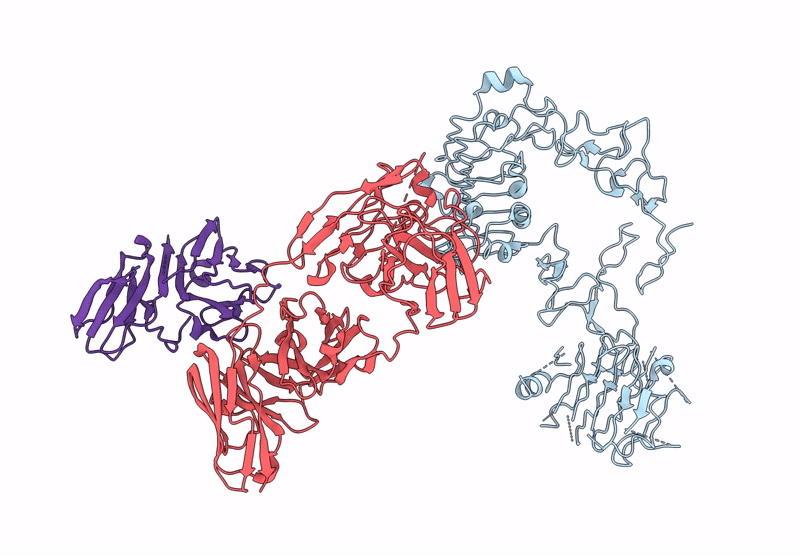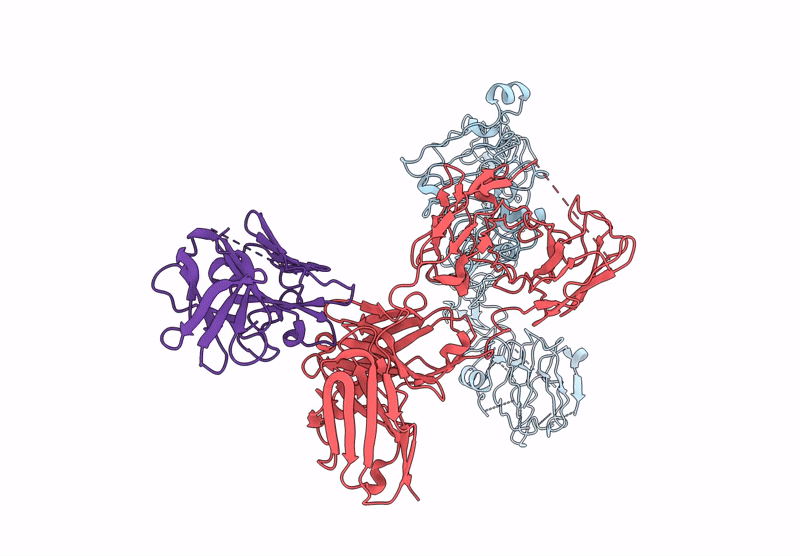
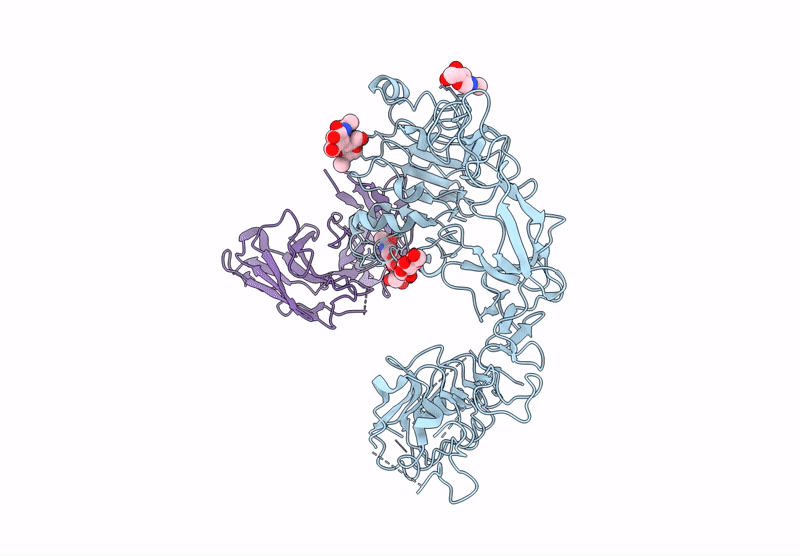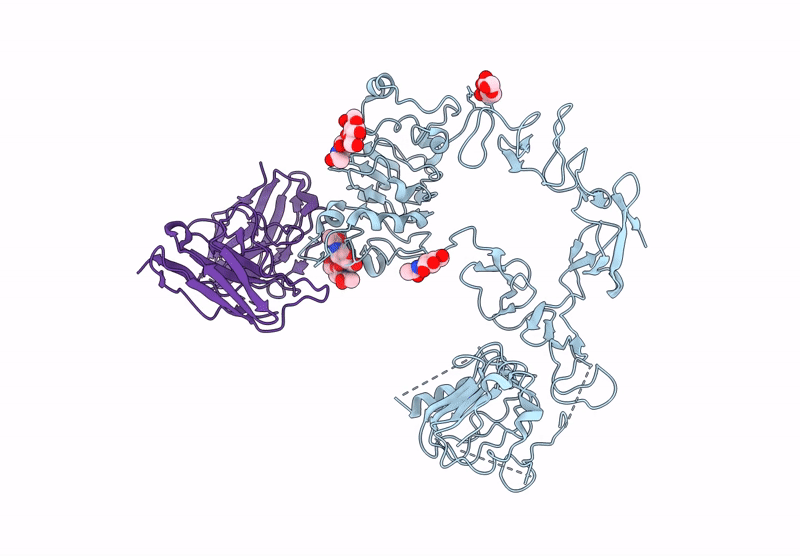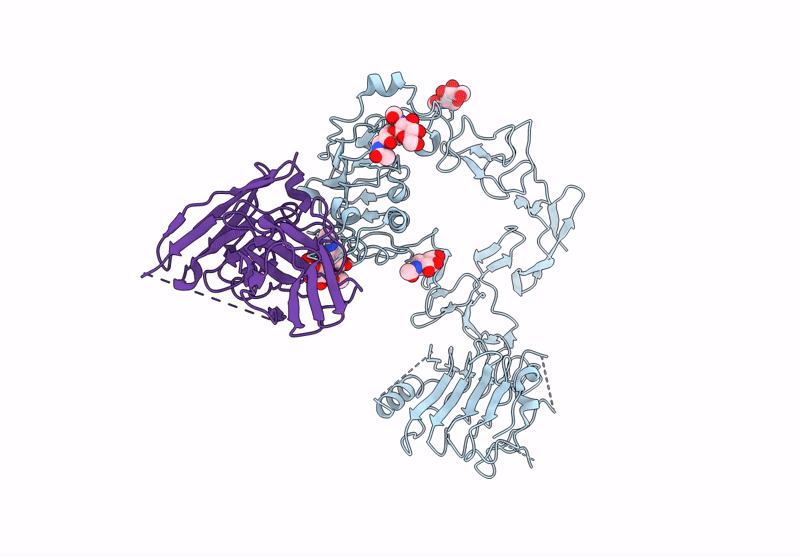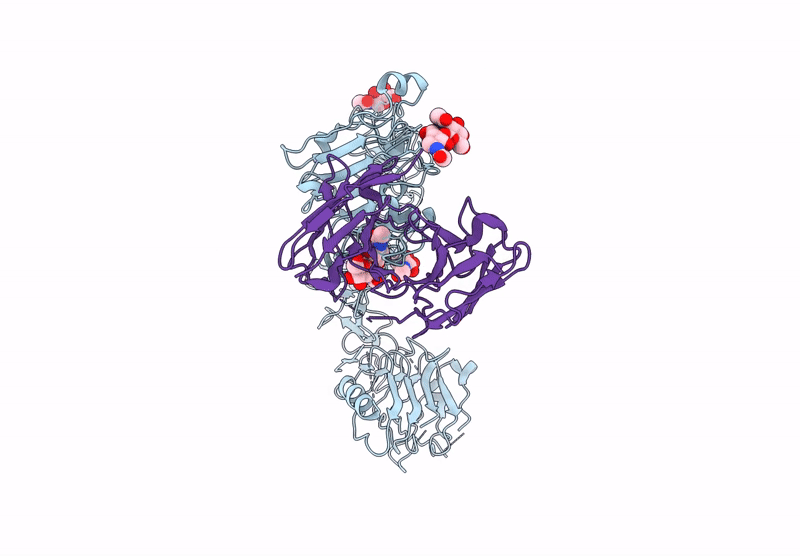
Local Refinement Structure Of Segfr And 528 Fv (From Hl-Type Bispecific Diabody Ex3) Complex
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Poly-Alanine Model For Hl-Type Bispecific Diabody Ex3 Composed Of 528 And Okt3 Fvs In Ternary Complex With Segfr And Cd3Gamma-Epsilon (Closed Conformation)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
 |
Poly-Alanine Model For Hl-Type Bispecific Diabody Ex3 Composed Of 528 And Okt3 Fvs In Ternary Complex With Segfr And Cd3Gamma-Epsilon (Middle Conformation)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
 |
Poly-Alanine Model For Hl-Type Bispecific Diabody Ex3 Composed Of 528 And Okt3 Fvs In Ternary Complex With Segfr And Cd3Gamma-Epsilon (Open Conformation)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
 |
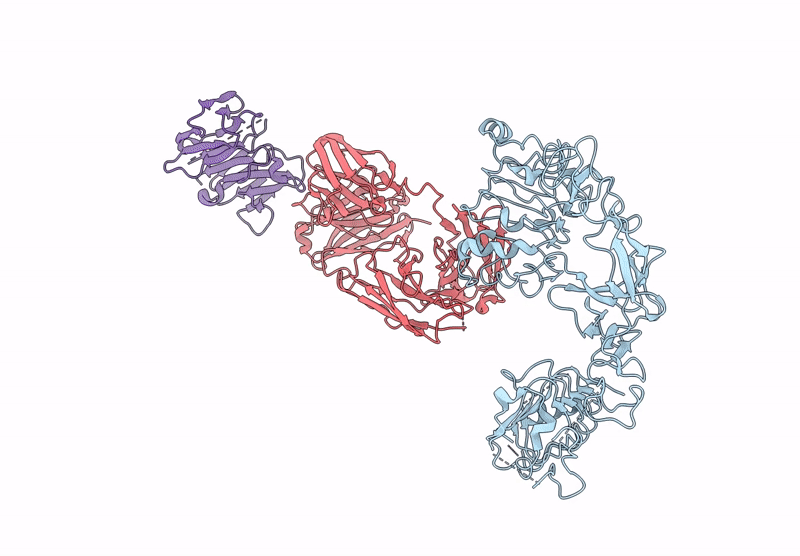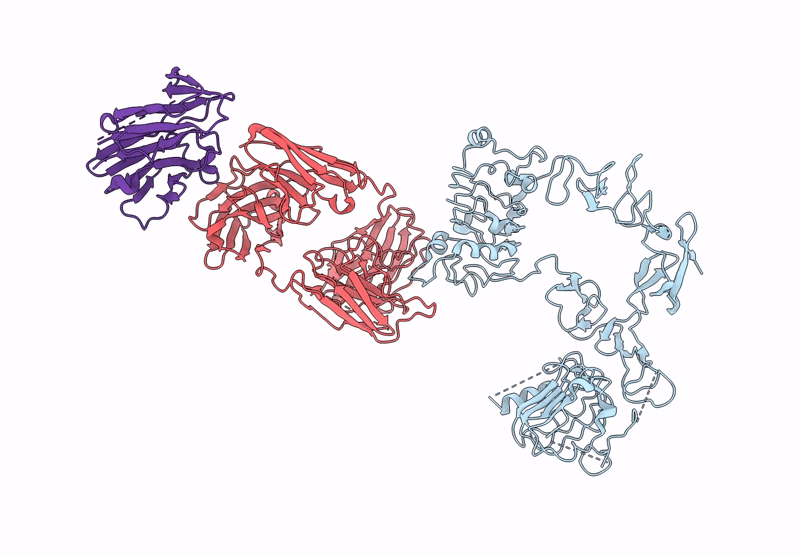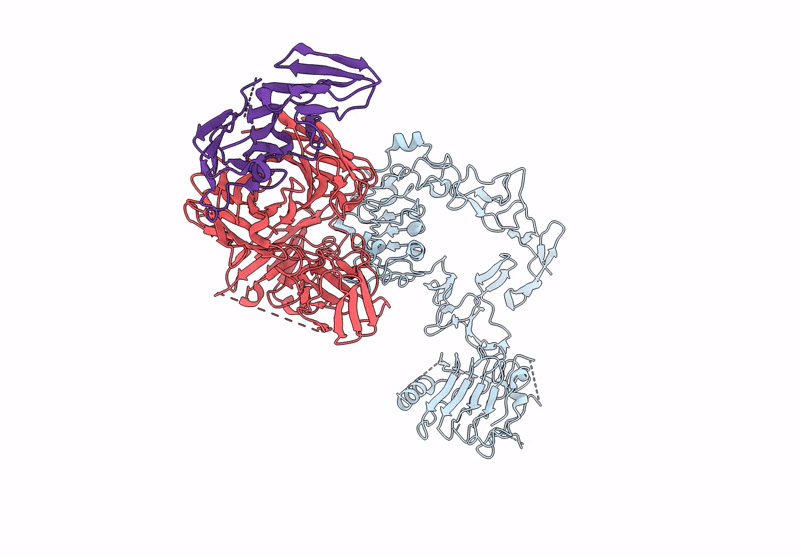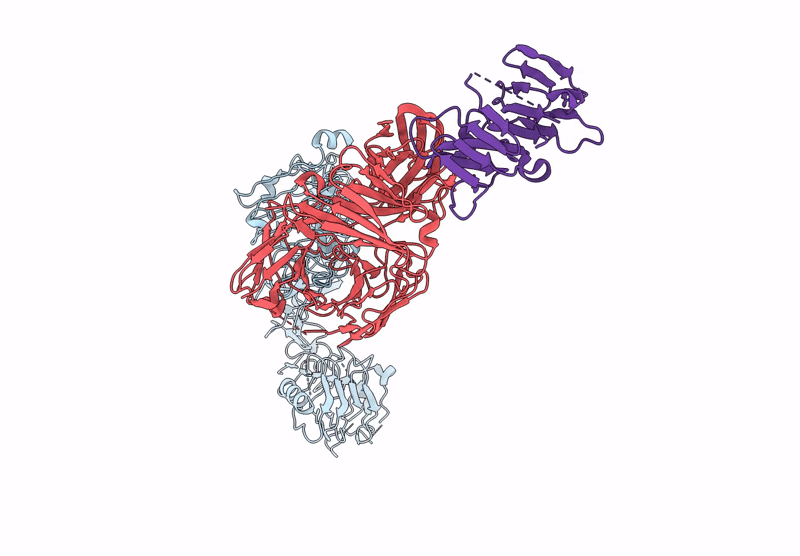
Local Refinement Structure Of Segfr And 528 Fv (From Lh-Type Bispecific Diabody Ex3) Complex
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Poly-Alanine Model For Lh-Type Bispecific Diabody Ex3 Composed Of 528 And Okt3 Fvs In Ternary Complex With Segfr And Cd3Gamma-Epsilon (Closed Conformation)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
 |
Poly-Alanine Model For Lh-Type Bispecific Diabody Ex3 Composed Of 528 And Okt3 Fvs In Ternary Complex With Segfr And Cd3Gamma-Epsilon (Middle Conformation)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
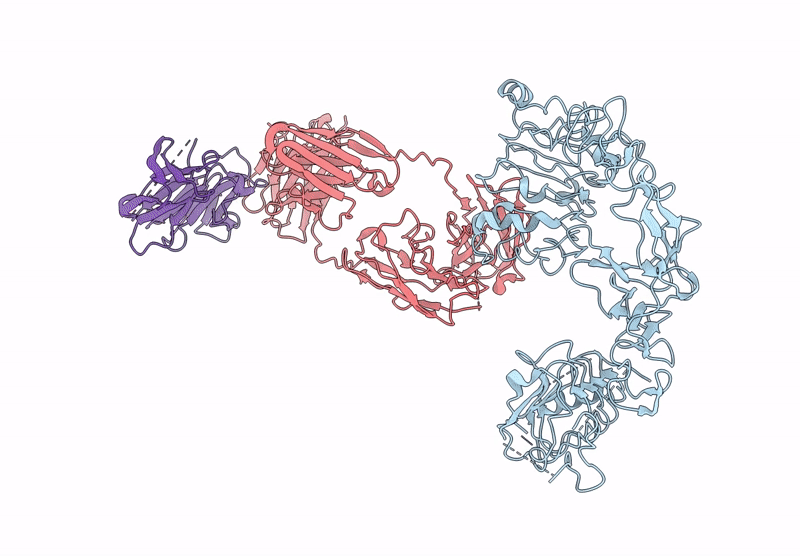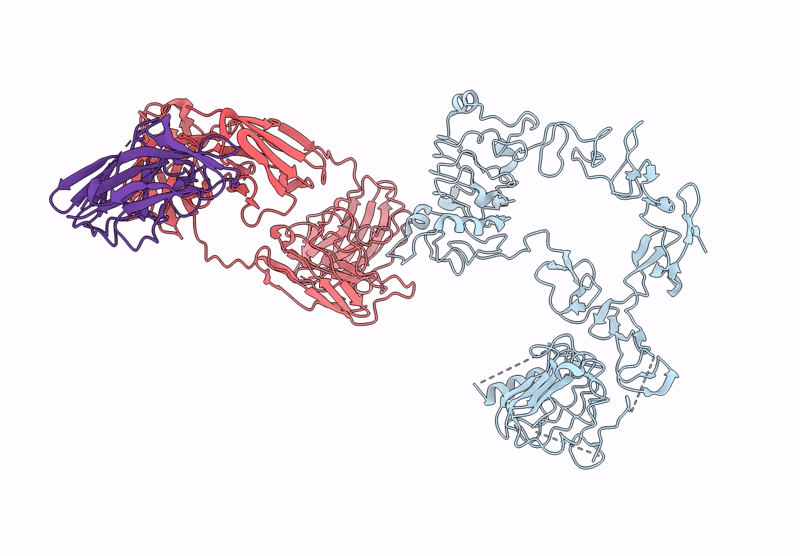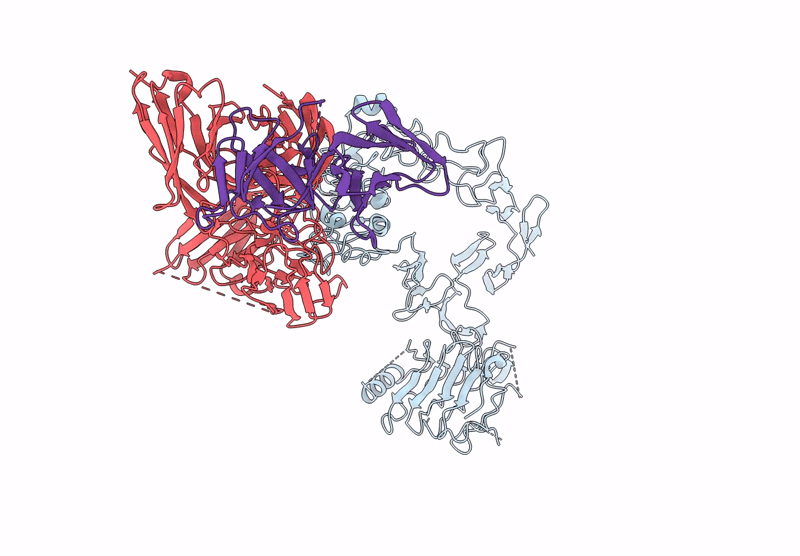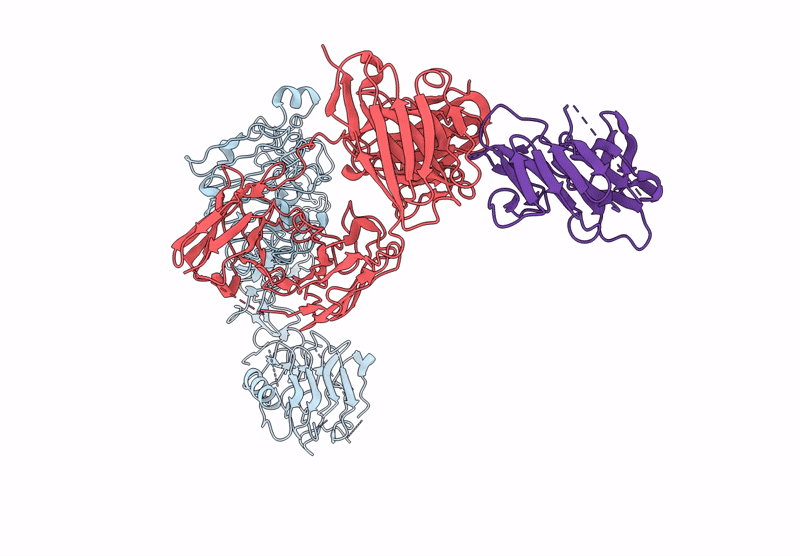 |
Poly-Alanine Model For Lh-Type Bispecific Diabody Ex3 Composed Of 528 And Okt3 Fvs In Ternary Complex With Segfr And Cd3Gamma-Epsilon (Open Conformation)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
 |
Organism: Streptomyces griseoluteus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-04-27 Classification: TRANSCRIPTION |
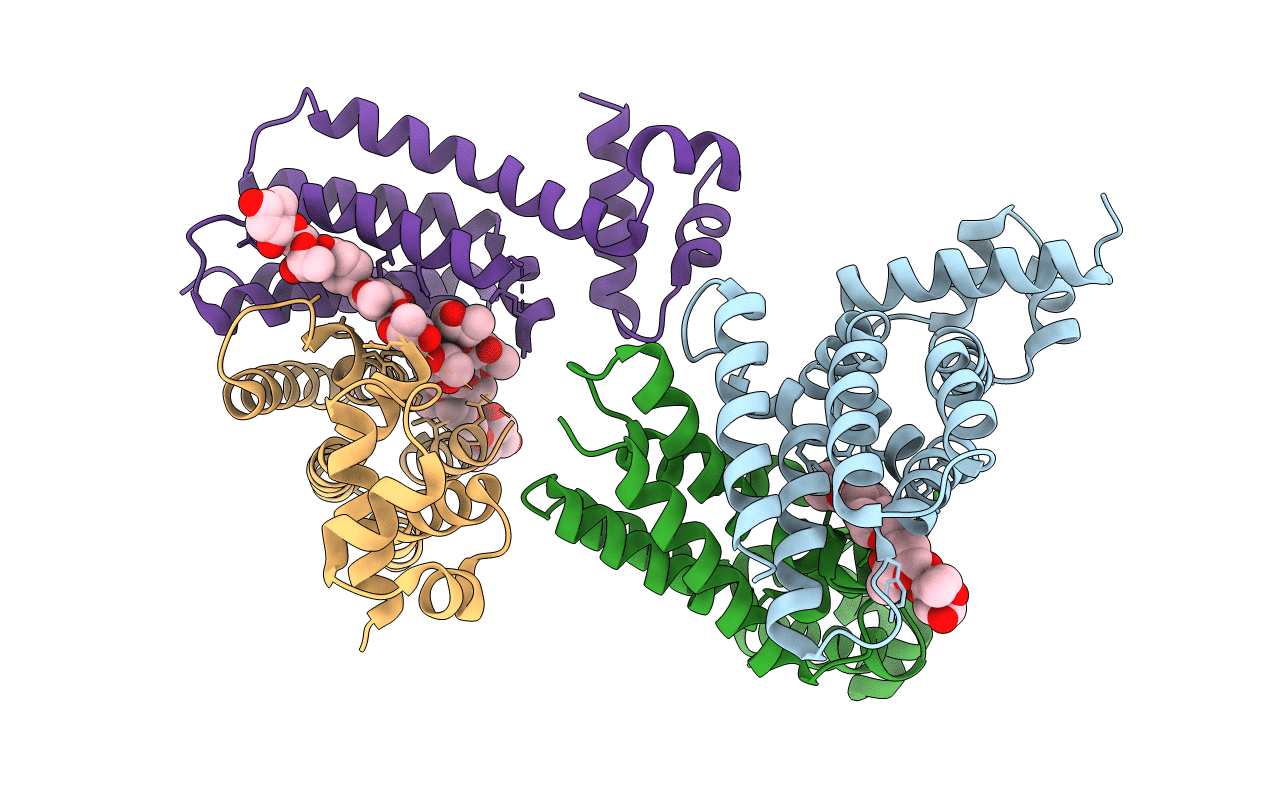 |
Organism: Streptomyces griseoluteus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2022-04-27 Classification: TRANSCRIPTION Ligands: JB0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-12-05 Classification: TRANSFERASE |
 |
Crystal Structure Of The Human Cap-Specific Adenosine Methyltransferase Bound To Sah
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH |
 |
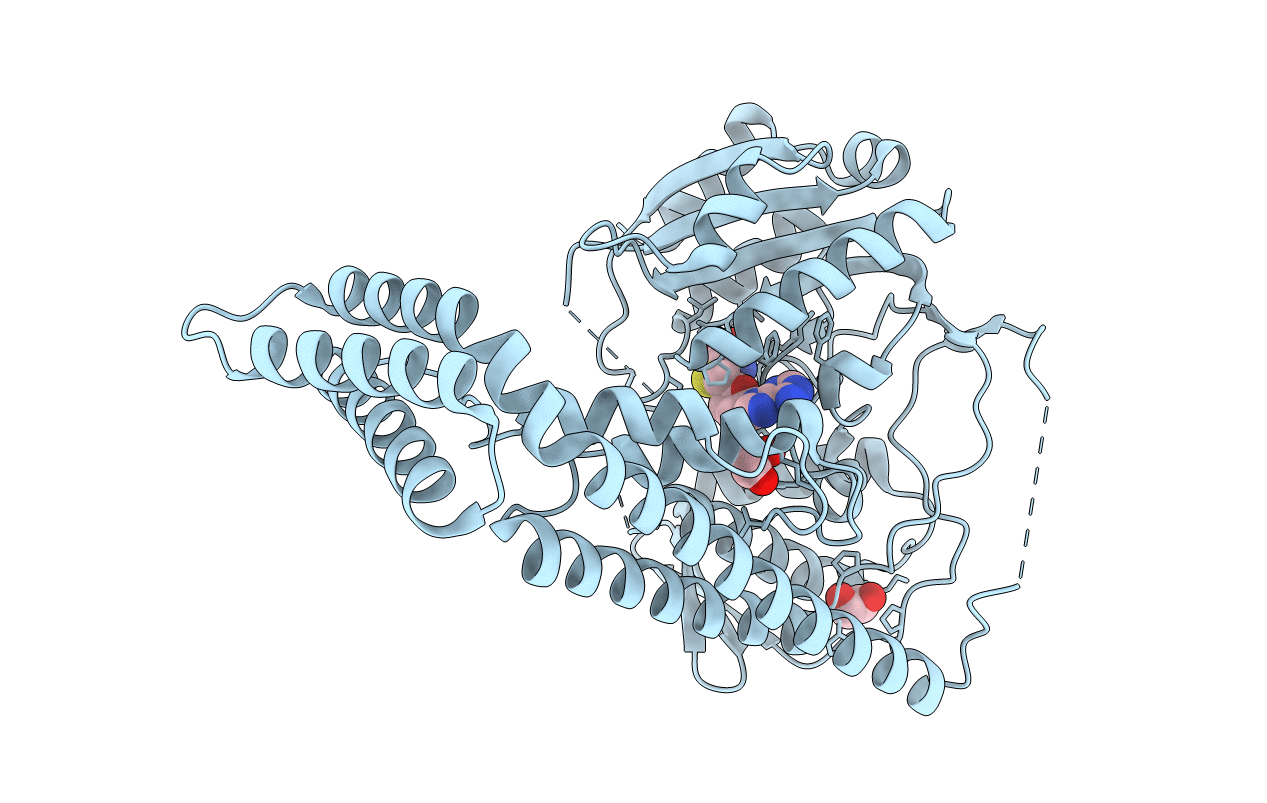
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-05 Classification: TRANSFERASE |
 |
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase Bound To Sah
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH, EDO |
 |
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase Bound To Sah And M7G-Capped Rna
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH, EDO, M7G |
 |
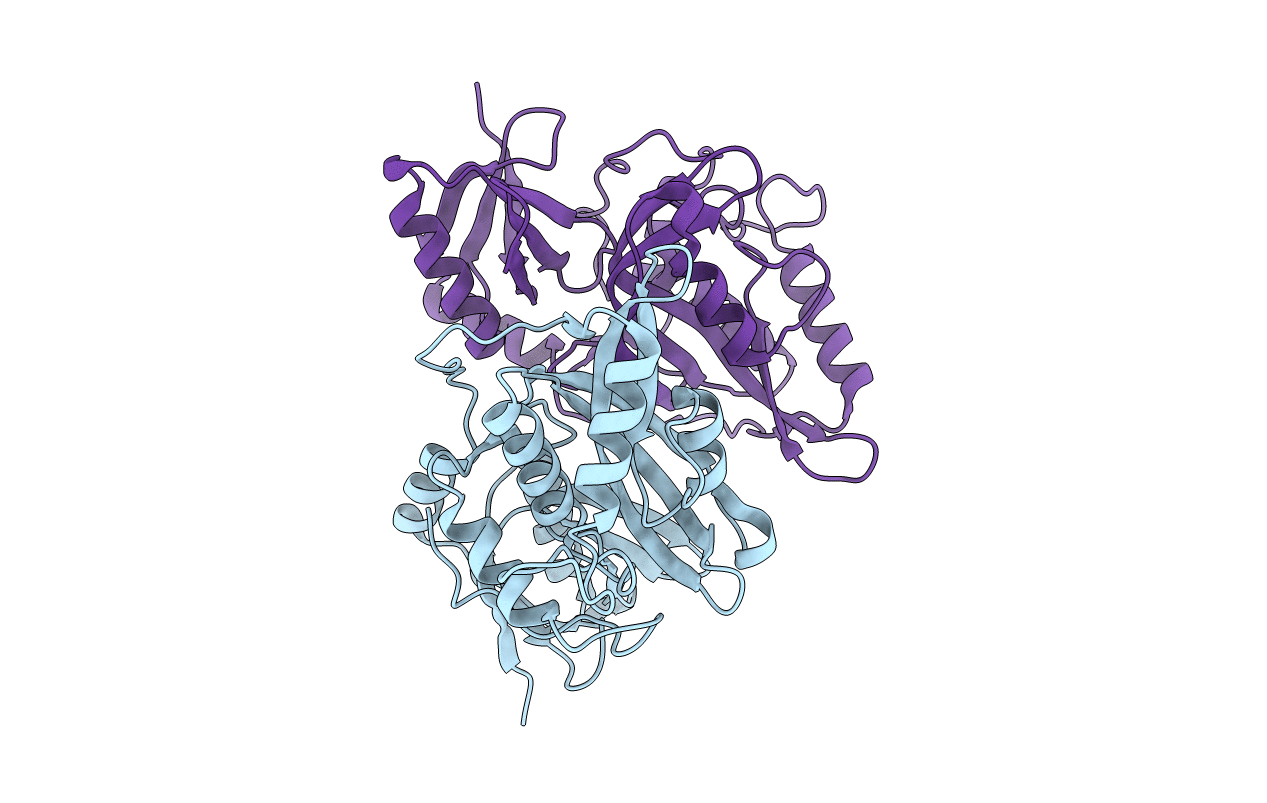
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase Bound To Sah And M7G-Capped Rna
Organism: Danio rerio, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-12-05 Classification: TRANSFERASE/RNA Ligands: SAH, M7G, EDO, B3P |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2010-08-11 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2009-03-17 Classification: HYDROLASE |

