Search Count: 43
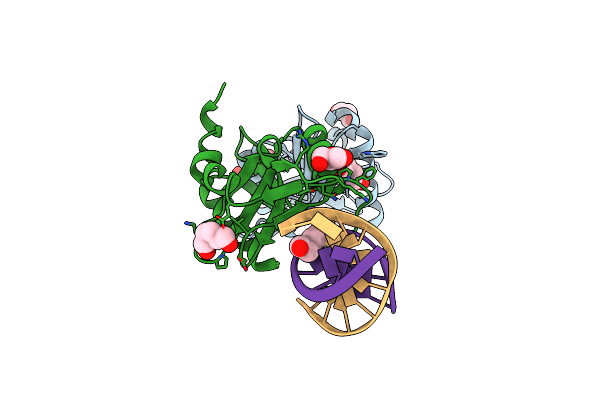 |
Crystal Structures Of Holliday Junction Resolvases From Archaeoglobus Fulgidus Bound To Dna Substrate
Organism: Archaeoglobus fulgidus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-05-26 Classification: HYDROLASE/DNA Ligands: HEZ, GOL |
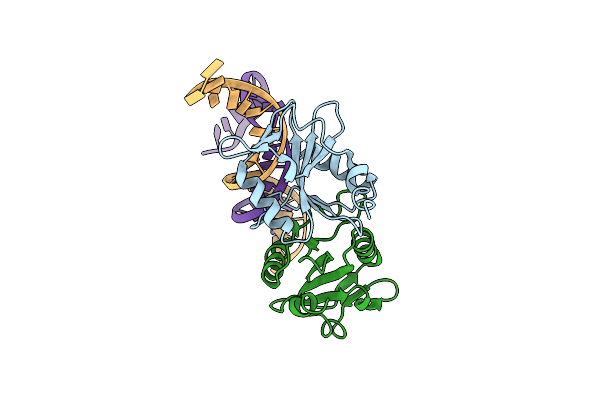 |
Crystal Structures Of Holliday Junction Resolvases From Archaeoglobus Fulgidus Bound To Dna Substrate
Organism: Archaeoglobus fulgidus dsm 4304, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2009-05-26 Classification: HYDROLASE/DNA |
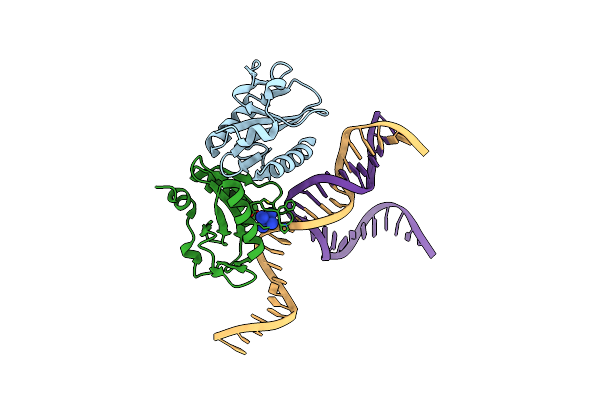 |
Crystal Structures Of Holliday Junction Resolvases From Archaeoglobus Fulgidus Bound To Dna Substrate
Organism: Archaeoglobus fulgidus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2009-05-26 Classification: HYDROLASE/DNA Ligands: NCO |
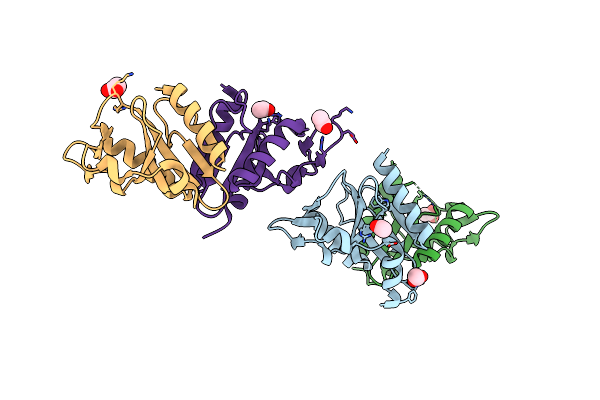 |
1.6A Resolution Structure Of Archaeoglobus Fulgidus Hjc, A Holliday Junction Resolvase From An Archaeal Hyperthermophile
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2009-03-24 Classification: HYDROLASE Ligands: ACT, NH4 |
 |
1.6A Resolution Structure Of Archaeoglobus Fulgidus Hjc, A Holliday Junction Resolvase From An Archaeal Hyperthermophile
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-03-24 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of T4 Endonuclease Vii N62D Mutant In Complex With A Dna Holliday Junction
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2008-01-29 Classification: Hydrolase/DNA Ligands: MG, ZN, EDO |
 |
Crystal Structure Of T4 Endonuclease Vii H43N Mutant In Complex With Heteroduplex Dna Containing Base Mismatches
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-01-29 Classification: Hydrolase/DNA Ligands: ZN |
 |
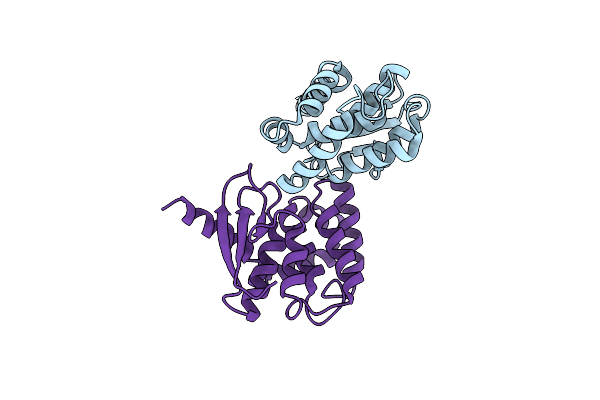
Crystal Structures Of The Interacting Domains From Yeast Glutamyl-Trna Synthetase And Trna Aminoacylation And Nuclear Export Cofactor Arc1P Reveal A Novel Function For An Old Fold
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-09-05 Classification: BIOSYNTHETIC PROTEIN, RNA BINDING Ligands: SO4 |
 |
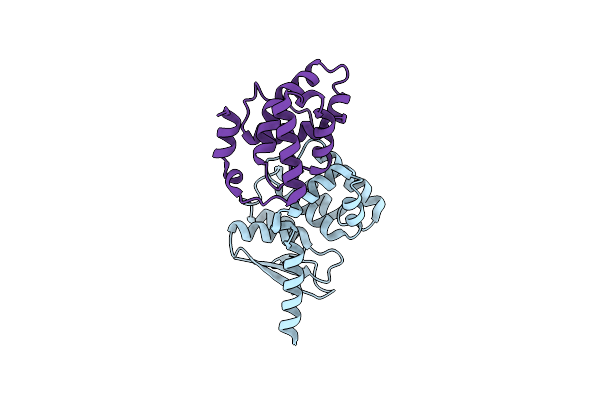
Crystal Structures Of The Interacting Domains From Yeast Glutamyl-Trna Synthetase And Trna Aminoacylation And Nuclear Export Cofactor Arc1P Reveal A Novel Function For An Old Fold
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-09-05 Classification: LIGASE Ligands: IOD |
 |
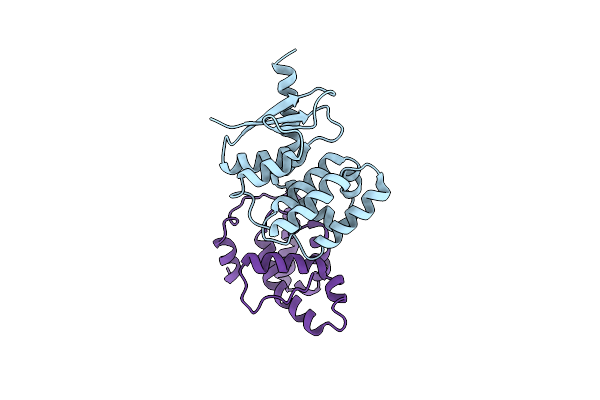
Structural Basis Of Yeast Aminoacyl-Trna Synthetase Complex Formation Revealed By Crystal Structures Of Two Binary Sub-Complexes
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2006-09-05 Classification: ligase/RNA Binding Protein Ligands: CL |
 |
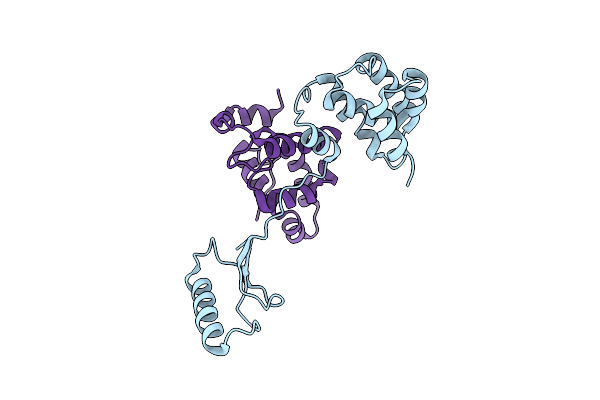
Structural Basis Of Yeast Aminoacyl-Trna Synthetase Complex Formation Revealed By Crystal Structures Of Two Binary Sub-Complexes
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2006-09-05 Classification: Ligase/RNA Binding Protein |
 |
Structural Basis Of Yeast Aminoacyl-Trna Synthetase Complex Formation Revealed By Crystal Structures Of Two Binary Sub-Complexes
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-09-05 Classification: Ligase/RNA Binding Protein |
 |
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2005-06-07 Classification: RNA BINDING PROTEIN |
 |
Organism: Bacteriophage t5
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2004-02-05 Classification: HYDROLASE Ligands: MN |
 |
Organism: Bacteriophage t5
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2004-02-05 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-11-20 Classification: HYDROLASE Ligands: CA, XE |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2003-09-16 Classification: REPLICATION/DNA Ligands: MG, IOD |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2003-09-16 Classification: REPLICATION/DNA Ligands: IOD, MG |
 |
Crystal Structure Of A Wild-Type Cre Recombinase-Loxp Synapse: Pre-Cleavage Complex
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2003-09-16 Classification: REPLICATION/DNA Ligands: IOD, MG |
 |
Crystal Structure Of A Wild-Type Cre Recombinase-Loxp Synapse: Phosphotyrosine Covalent Intermediate
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2003-09-16 Classification: REPLICATION/DNA Ligands: IOD, MG |

