Search Count: 7
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-06-03 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN, GOL, BR |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.9000 Å, 2.2220 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, DOD |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.9000 Å, 2.0900 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, K, SO4, ASC, DOD |
 |
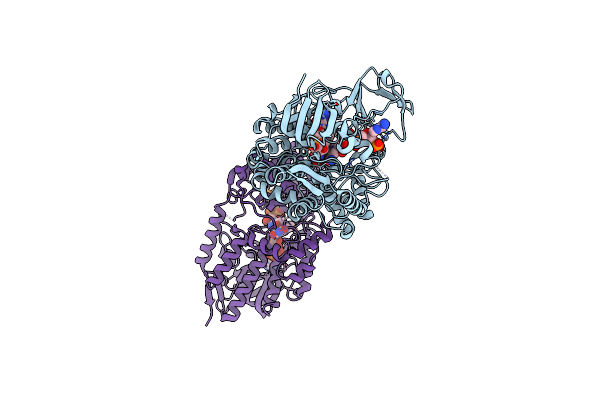
Crystal Structure Of Cyclohexanone Monooxygenase From Rhodococcus Sp. Phi1 Bound To Nadp+
Organism: Rhodococcus sp. phi1
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2018-09-26 Classification: FLAVOPROTEIN Ligands: FAD, NAP |
 |
Crystal Structure Of Cyclohexanone Monooxygenase Mutant (F249A, F280A And F435A) From Rhodococcus Sp. Phi1 Bound To Nadp+
Organism: Rhodococcus sp. phi1
Method: X-RAY DIFFRACTION Release Date: 2018-09-26 Classification: FLAVOPROTEIN Ligands: FAD, NAP |
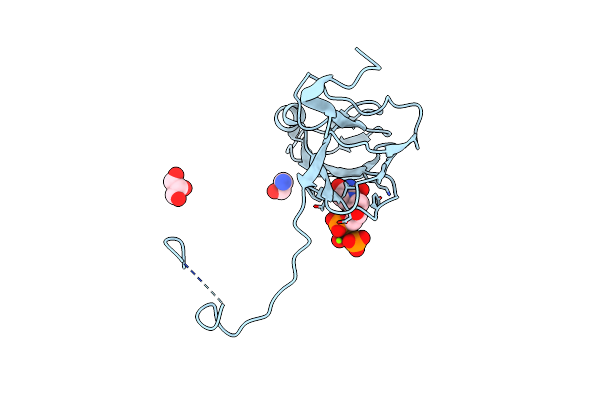 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: DUP, TRS, MG, GOL |
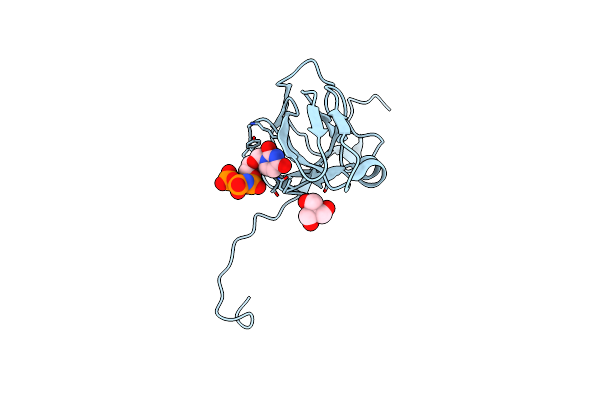 |
Crystal Structure Of Mycobacterium Tuberculosis Dutpase R140K, H145W Mutant
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: TRS, DUP, MG |

