Search Count: 18
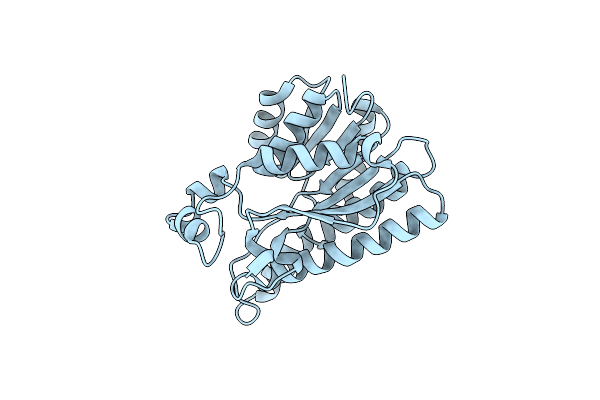 |
X-Ray Structure Of 3-Alpha-(Or 20-Beta)-Hydroxysteroid Dehydrogenase Mutant
Organism: Lactobacillaceae
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE |
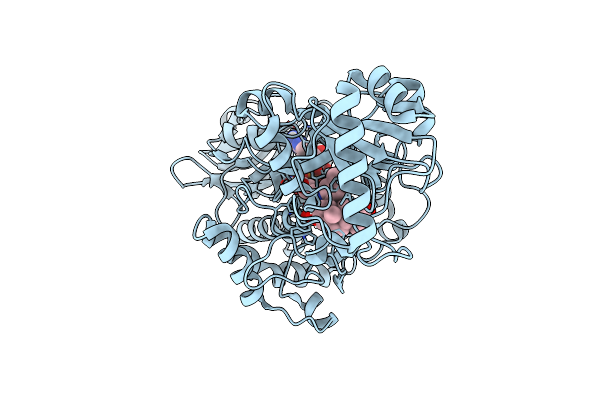 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 1,4-Androstadiene-3,17- Dione
Organism: Rhodococcus electrodiphilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD, ANB |
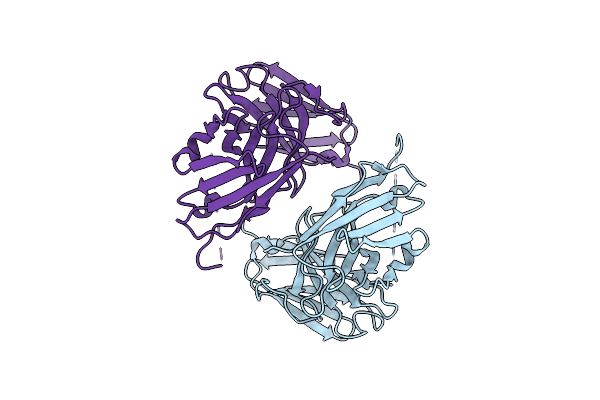 |
Crystal Structure Of Single-Chain Fv Antibody Against Antigen Peptide From Sars-Cov2 S-Spike Protein
Organism: Escherichia coli (strain k12), Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-11-08 Classification: IMMUNE SYSTEM |
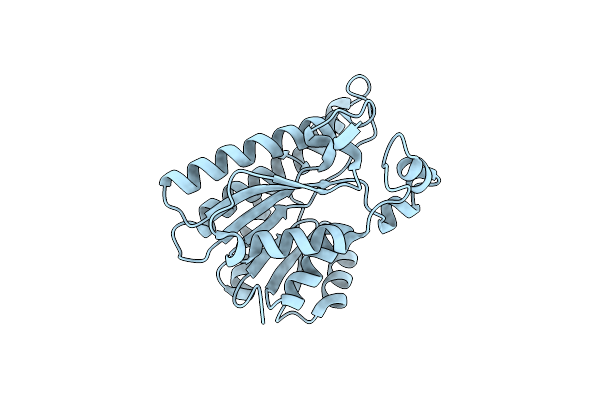 |
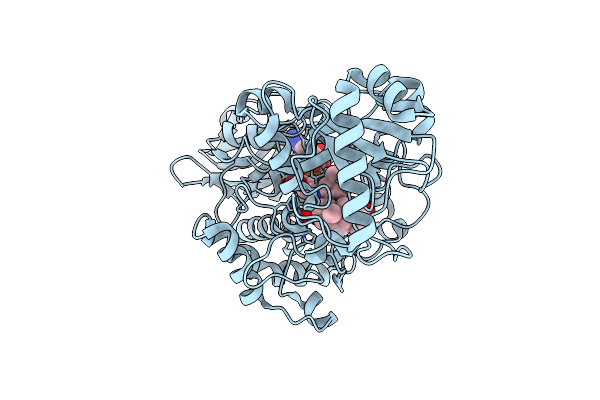
Organism: Lentilactobacillus kefiri
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 1,4-Androstadiene-3,17- Dione
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, ANB |
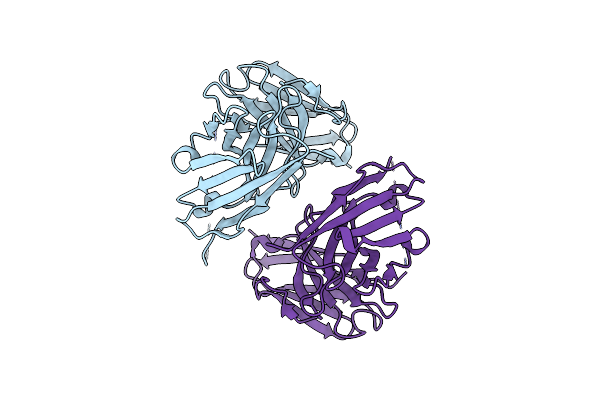 |
Crystal Structure Of Scfv Against The Receptor Binding Domine Of Sars-Cov-2 S-Spike Protein
Organism: Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-08-23 Classification: IMMUNE SYSTEM |
 |
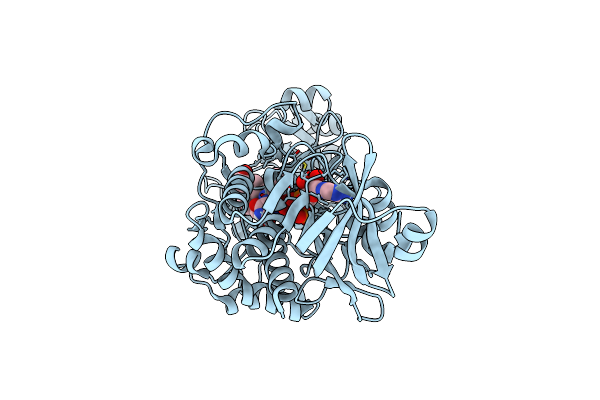
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 9,11-Epoxy-17-Hydroxypregn-4-Ene-3,20-Dione Actate
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: FAD, UU0 |
 |
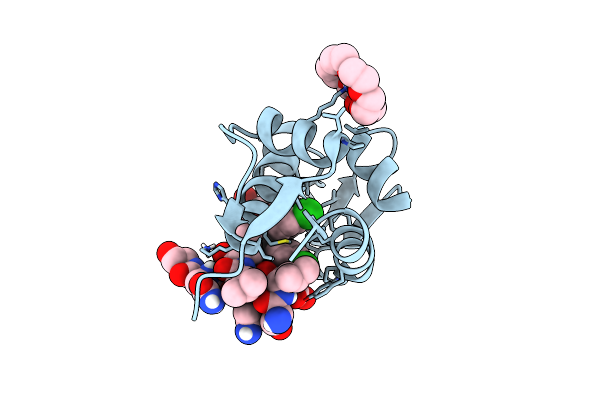
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 4-Androstadiene-3,17- Dione
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, ASD |
 |
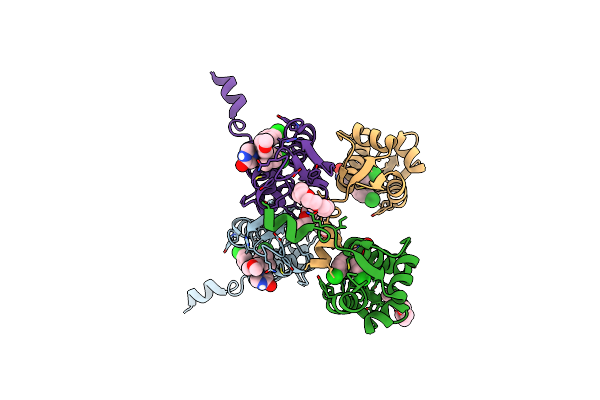
Small Peptide Enhances The Binding Of Nutline-3A To N-Terminal Domain Of Mdmx
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-02-22 Classification: ONCOPROTEIN Ligands: NUT, O4B |
 |
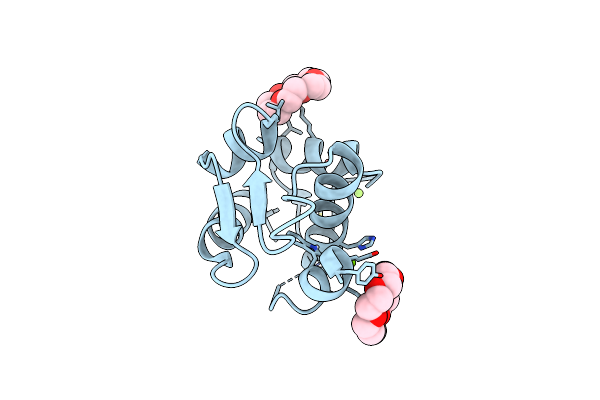
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-11-30 Classification: ONCOPROTEIN Ligands: NUT, O4B |
 |
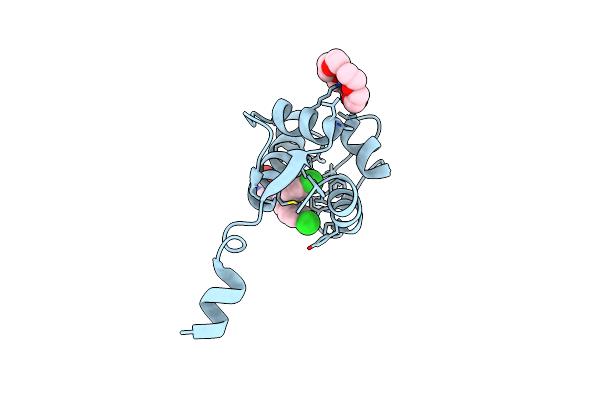
The Crystal Structure Of P53P Peptide Fragment In Complex With The N-Terminal Domain Of Mdmx
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2021-06-09 Classification: ONCOPROTEIN Ligands: O4B, MG |
 |
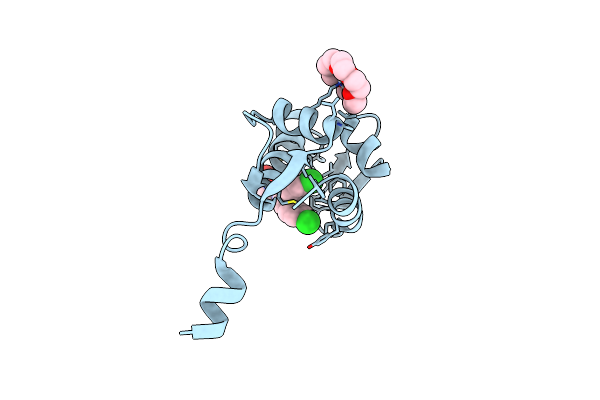
Crystal Structure Of The N-Terminal Domain Of Human Mdmx Protein In Complex With Nutlin3A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2020-06-17 Classification: ONCOPROTEIN Ligands: O4B, NUT |
 |
Crystal Structure Of The P53-Binding Domain Of Human Mdmx Protein In Complex With Nutlin3A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-06-17 Classification: ONCOPROTEIN Ligands: NUT, O4B |
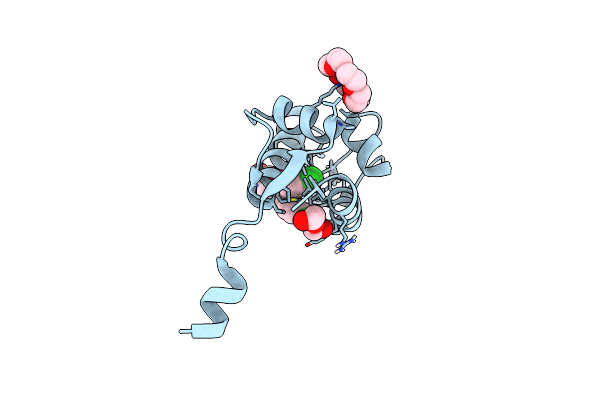 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-06-10 Classification: ONCOPROTEIN Ligands: O4B, NUT, PEG |
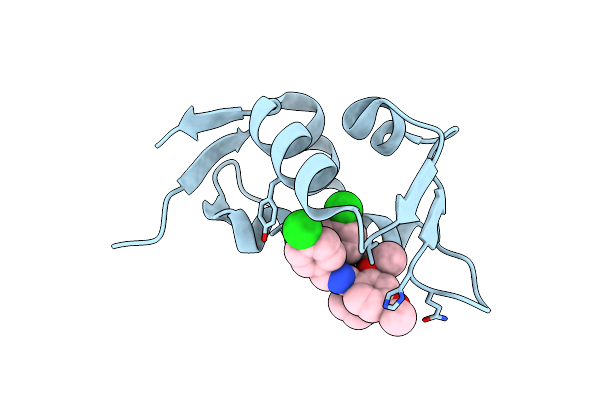 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2018-08-22 Classification: CELL CYCLE Ligands: NUT |
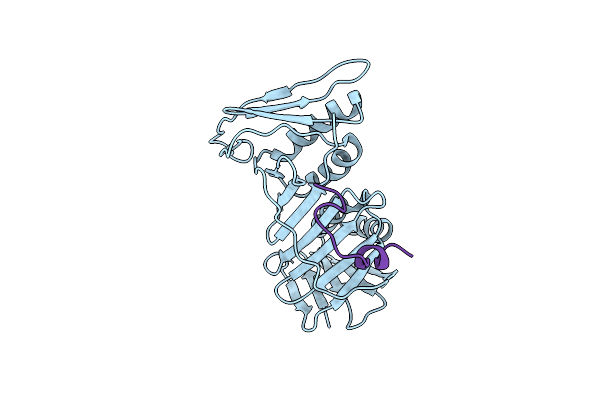 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2018-05-30 Classification: CELL CYCLE |
 |
Crystal Structure Of Delta 5-3-Ketosteroid Isomerase From Mycobacterium Sp.
Organism: Mycobacterium neoaurum
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2018-01-31 Classification: BIOSYNTHETIC PROTEIN |
 |
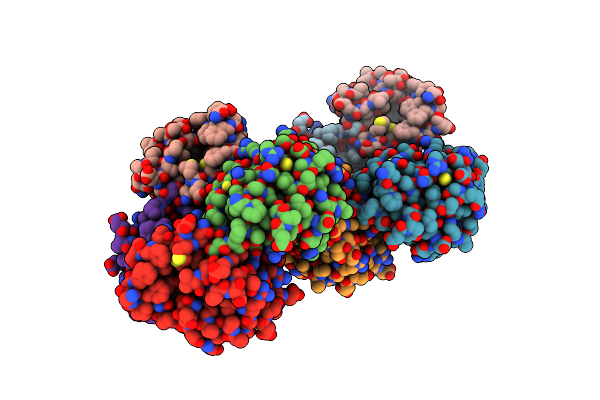
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2018-01-03 Classification: ONCOPROTEIN Ligands: NUT |

