Search Count: 312
 |
Organism: Tribonema minus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: PHOTOSYNTHESIS Ligands: CLA, BCR, DD6, LMT, LHG, A1L1G, LMG, PQN, SF4 |
 |
Organism: Adeno-associated virus - 8, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Adeno-associated virus - 8
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRUS |
 |
Organism: Adeno-associated virus - 8, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Henipavirus nipahense, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Pseudomonas aeruginosa pao1
Method: SOLUTION NMR Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: SIGNALING PROTEIN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: ENDOCYTOSIS Ligands: NAG, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: ENDOCYTOSIS Ligands: NAG, CA, HEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: ENDOCYTOSIS Ligands: NAG, CA, HEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: EXOCYTOSIS Ligands: NAG, CA, HEM |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2025-01-01 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, A1LX0, MG, TPP |
 |
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: PROTEIN TRANSPORT Ligands: ZN, DGA, DGD, LMG, ACD, SQD |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, ADP, ENO, MG |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, TPP, MG |
 |
Organism: Chryseomicrobium sp.
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, TPP, MG |
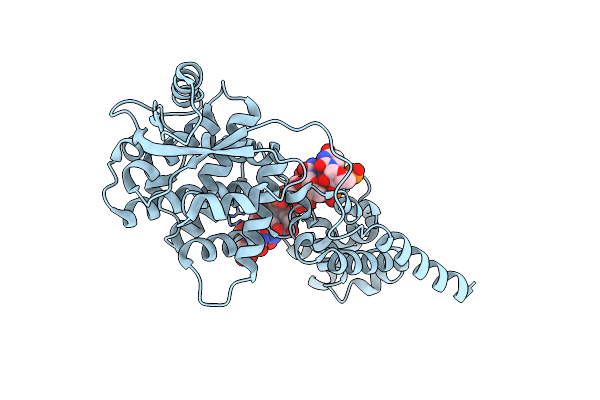 |
Organism: Mumps virus (strain jeryl-lynn), Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2024-08-21 Classification: NUCLEAR PROTEIN |
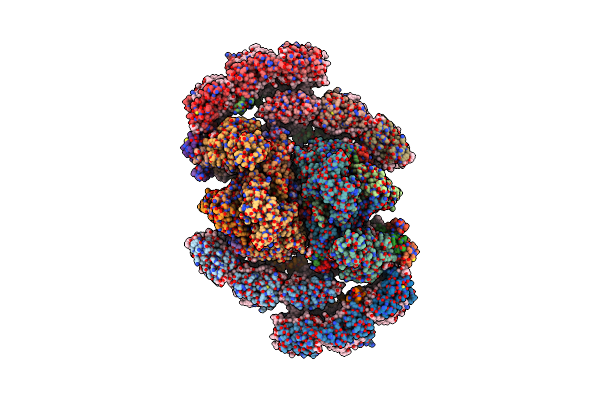 |
Structure Of Inactive Photosystem Ii Associated With Cac Antenna From Rhodomonas Salina
Organism: Rhodomonas salina
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: PHOTOSYNTHESIS Ligands: FE2, CLA, PHO, WVN, SQD, PL9, BCT, LHG, CL, MN, LMG, DGD, HEM, KC2, II0, IHT |
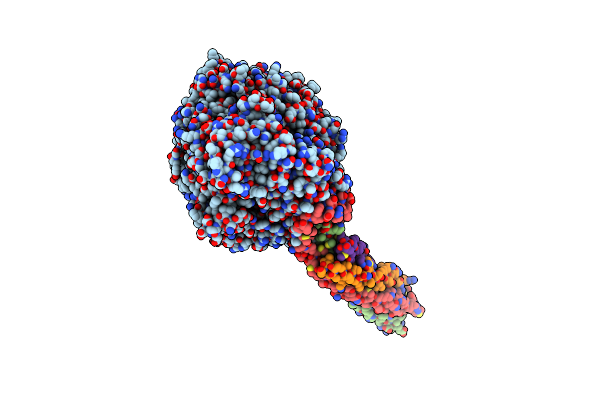 |
Structure Of The Mumps Virus L Protein (State2) Bound By Phosphoprotein Tetramer
Organism: Mumps orthorubulavirus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN Ligands: ZN |

