Search Count: 21
 |
Crystal Structure Of The Lon-Like Protease Mtalonc With D581A Mutation In Complex With Substrate Polypeptide
Organism: Meiothermus taiwanensis, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of The Lon-Like Protease Mtalonc With D582A Mutation In Complex With Substrate Polypeptide
Organism: Meiothermus taiwanensis, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of The Lon-Like Protease Mtalonc With S582A Mutation In Complex With F-B20-Q
Organism: Meiothermus taiwanensis, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of The Lon-Like Protease Mtalonc With D581A Mutation In Complex With F-B20-Q
Organism: Meiothermus taiwanensis, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-11-24 Classification: HYDROLASE Ligands: PO4 |
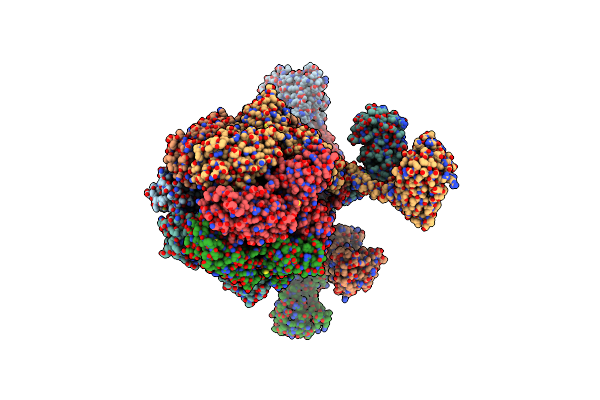 |
Processive Cleavage Of Substrate At Individual Proteolytic Active Sites Of The Lon Proteasecomplex (Conformation 1)
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: HYDROLASE Ligands: AGS, ADP |
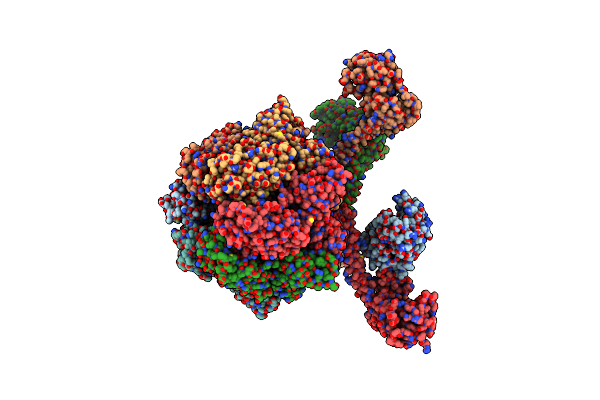 |
Processive Cleavage Of Substrate At Individual Proteolytic Active Sites Of The Lon Protease Complex (Conformation 2)
Organism: Meiothermus taiwanensis, Meiothermus taiwanensis wr-220
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: HYDROLASE Ligands: AGS, ADP |
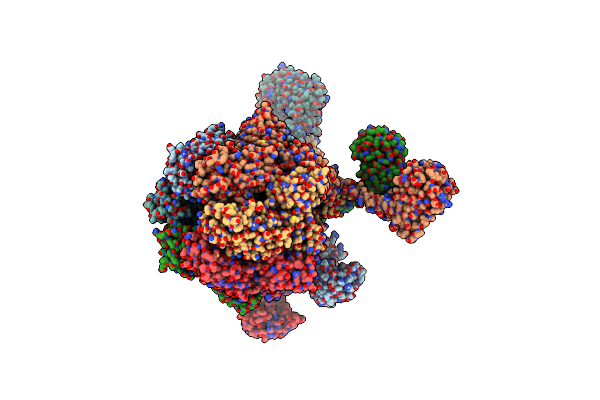 |
Processive Cleavage Of Substrate At Individual Proteolytic Active Sites Of The Lon Protease Complex (Conformation 3)
Organism: Meiothermus taiwanensis, Meiothermus taiwanensis wr-220
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: HYDROLASE Ligands: AGS, ADP |
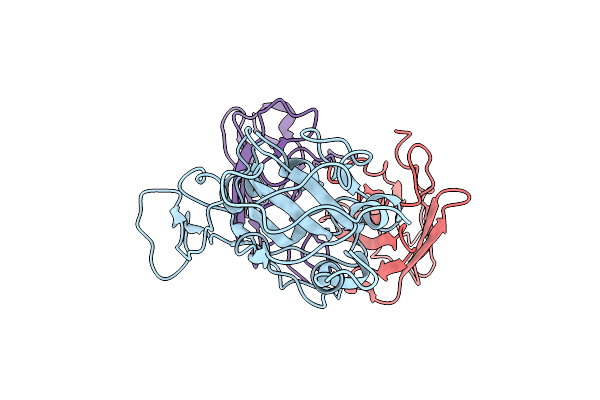 |
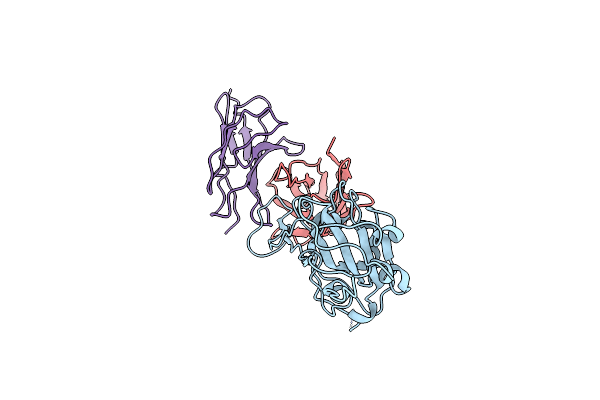
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With A Neutralizing Antibody Chab-25 (Focused Refinement Of S-Rbd And Chab-25 Region)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN |
 |
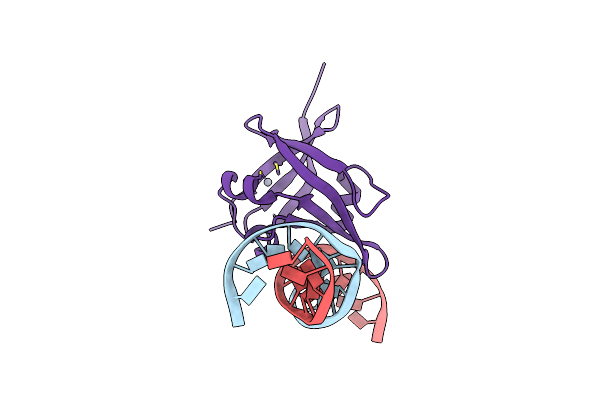
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With A Neutralizing Antibody Chab-45 (Focused Refinement Of S-Rbd And Chab-45 Region)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN |
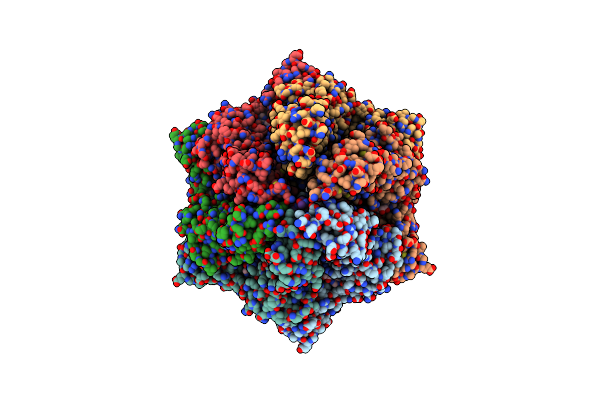 |
Molecular Basis For The Atpase-Powered Substrate Translocation By The Lon Aaa+ Protease
Organism: Meiothermus taiwanensis, Dictyostelium discoideum
Method: ELECTRON MICROSCOPY Release Date: 2021-06-09 Classification: MOTOR PROTEIN Ligands: AGS, 4KZ, ADP |
 |
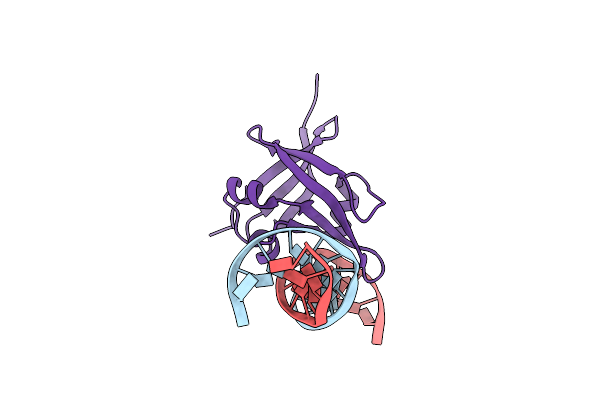
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2018-05-02 Classification: TRANSCRIPTION/DNA Ligands: HG |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2018-05-02 Classification: TRANSCRIPTION/DNA |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2018-05-02 Classification: TRANSCRIPTION/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: FBP, SER, PYR, MG, K |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-22 Classification: PROTEIN BINDING/SIGNALING PROTEIN/HYDROLASE/PEPTIDE |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-02-08 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2016-11-09 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2016-03-09 Classification: TRANSFERASE Ligands: PYR, MG, SER, FBP |
 |
Organism: Schizosaccharomyces pombe 972h-, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2016-01-27 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2016-01-27 Classification: RNA BINDING PROTEIN |

