Search Count: 125
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: HVG, A1ENS |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: GLU |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: Z99 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: A1ENR, HVG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: TCA, NA, EDO, PEG, PG4, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: A1A1H |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: A1A1H, PEG, GOL, EDO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: ZN, A1A1I |
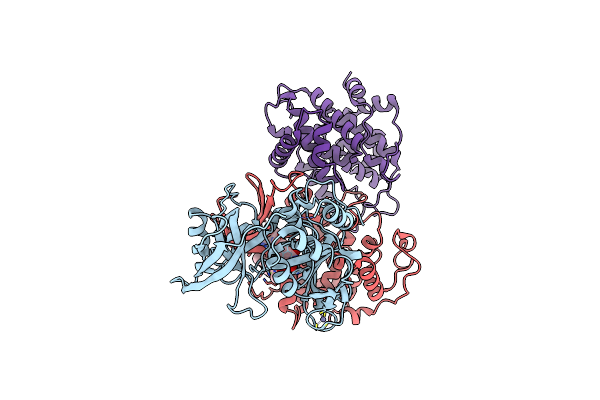 |
Cryo-Em Structure Of Cdk2/Cycline1 In Complex With Crbn/Ddb1 And Cpd 4 (Local Mask)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: ZN, A1A1I |
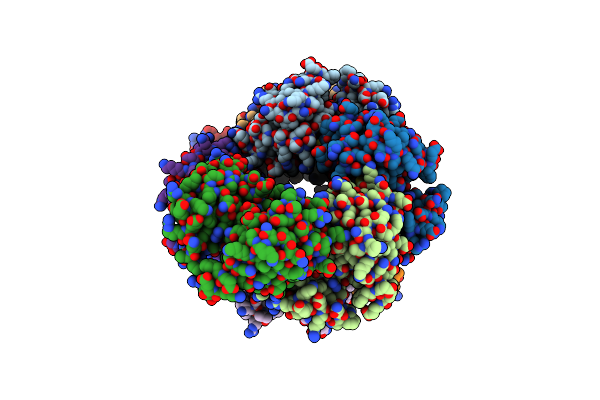 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-10-30 Classification: APOPTOSIS Ligands: SE |
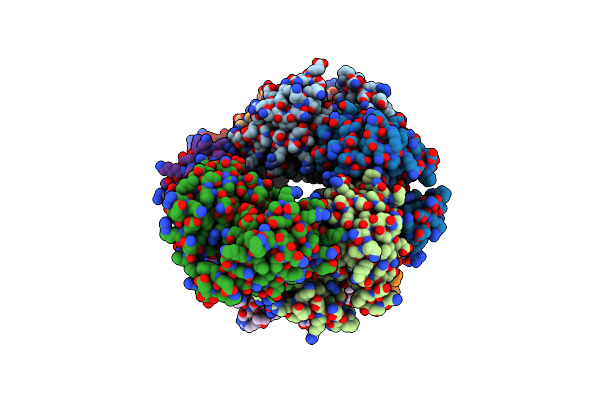 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-10-30 Classification: APOPTOSIS |
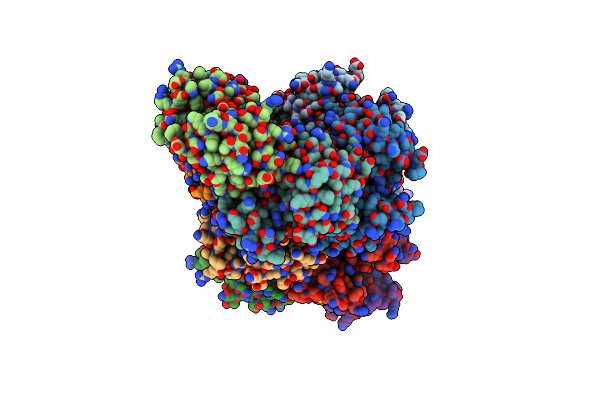 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-10-30 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: APOPTOSIS |
 |
Organism: Xenorhabdus doucetiae frm16 = dsm 17909
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-16 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Xenorhabdus doucetiae frm16 = dsm 17909
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-16 Classification: TRANSFERASE Ligands: SAH, GOL |

